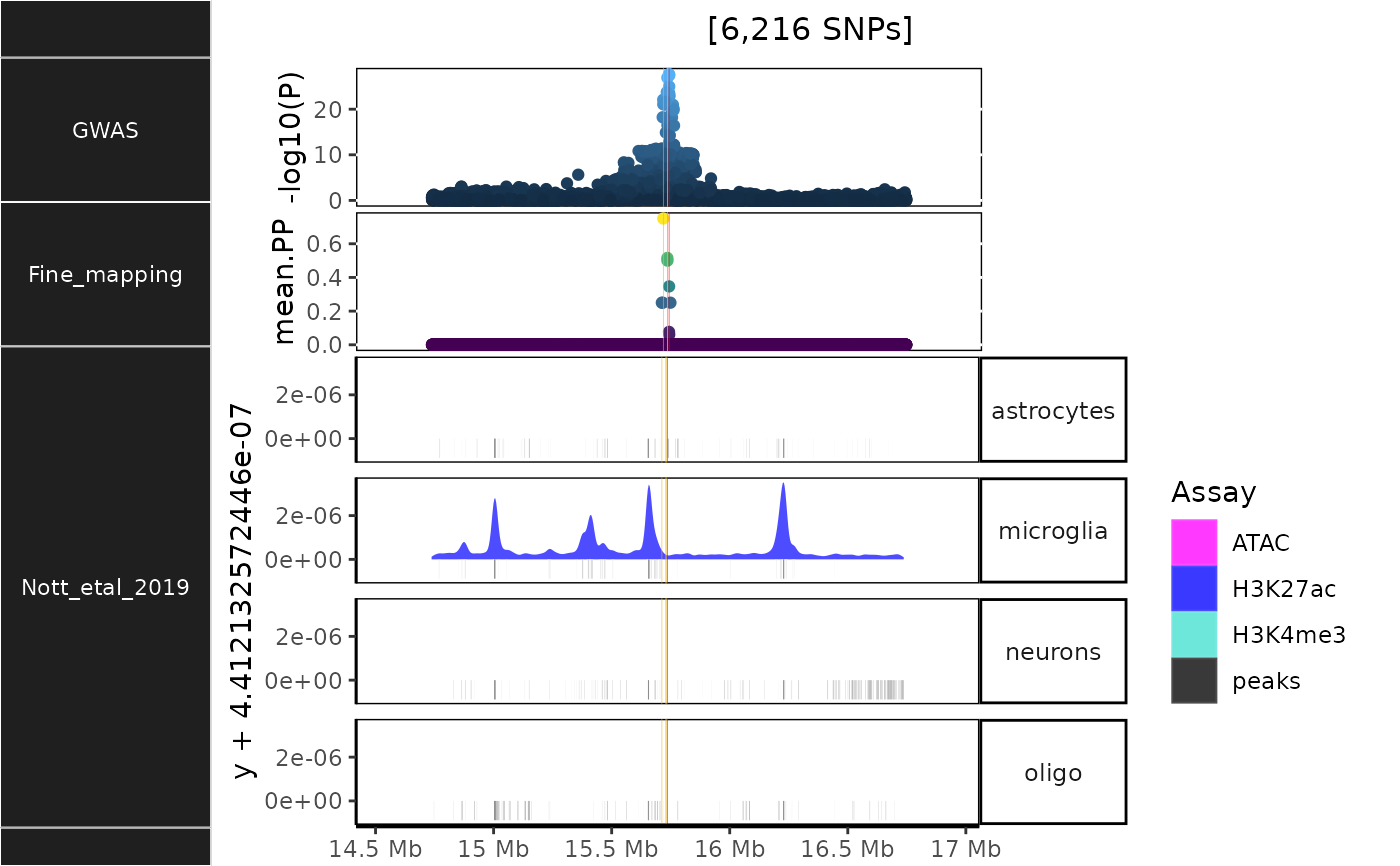
Plot brain cell-specific epigenomic data
Source:R/NOTT2019_epigenomic_histograms.R
NOTT2019_epigenomic_histograms.RdBrain cell-specific epigenomic data from Nott et al. (2019).
NOTT2019_epigenomic_histograms(
dat,
bigwig_metadata = echoannot::NOTT2019_bigwig_metadata,
locus_dir = tempdir(),
show_plot = TRUE,
save_plot = FALSE,
full_data = TRUE,
return_assay_track = FALSE,
binwidth = 200,
density_adjust = 0.2,
zoom = "1x",
strip.text.y.angle = 90,
xtext = TRUE,
geom = "density",
plot_formula = "Cell_type ~.",
fill_var = "Assay",
genomic_units = "Mb",
as_ggplot = TRUE,
dpi = 300,
height = 15,
width = 8,
nThread = 1,
save_annot = FALSE,
verbose = TRUE
)Source
Arguments
- dat
Fine-mapping results data from finemap_loci.
- bigwig_metadata
Metadata table with at least the following two columns:
"name"Unique name of the file.
"data_link"URL to UCSC genome browser bigwig file.
- locus_dir
Locus-specific directory.
- show_plot
Show plot.
- save_plot
Whether to save the plot.
- full_data
Whether to download the full data (genomic ranges of all sequence reads) as opposed to a reduced representation of the data as a single vector (i.e. the aggregated reads "score"). Setting
full_data=TRUEis necessary for creating histograms and density plots.- return_assay_track
Return only the assay track (before adding the rest of the tracks and showing the plot).
- binwidth
width of the bins.
- density_adjust
Passed to
adjustargument in geom_density.- zoom
Zoom into the center of the locus when plotting (without editing the fine-mapping results file). You can provide either:
The size of your plot window in terms of basepairs (e.g.
zoom=50000for a 50kb window).How much you want to zoom in (e.g.
zoom="1x"for the full locus,zoom="2x"for 2x zoom into the center of the locus, etc.).
You can pass a list of window sizes (e.g.
c(50000,100000,500000)) to automatically generate multiple views of each locus. This can even be a mix of different style inputs: e.g.c("1x","4.5x",25000).- strip.text.y.angle
Angle of the y-axis facet labels.
- xtext
Whether to include x-axis title and text.
- geom
Geom to use (Single character for now). Please see section Geometry for details.
- plot_formula
Formula passed to
facetsargument in facet_grid.- fill_var
Variable name to use for plot
fillargument.- genomic_units
Which genomic units to return window limits in.
- as_ggplot
Return plot as
ggplot2(TRUE) orTracks(FALSE) object.- dpi
dpi to use for raster graphics
- height
height (defaults to the height of current plotting window)
- width
width (defaults to the width of current plotting window)
- nThread
Number of threads to parallelise downloads across.
- save_annot
Save the queried subset of bigwig annotations.
- verbose
Print messages.
See also
Other NOTT2019:
NOTT2019_bigwig_metadata,
NOTT2019_get_epigenomic_peaks(),
NOTT2019_get_interactions(),
NOTT2019_get_interactome(),
NOTT2019_get_promoter_celltypes(),
NOTT2019_get_promoter_interactome_data(),
NOTT2019_get_regulatory_regions(),
NOTT2019_plac_seq_plot(),
NOTT2019_superenhancers(),
get_NOTT2019_interactome(),
get_NOTT2019_superenhancer_interactome()
Examples
nott2019_track <- echoannot::NOTT2019_epigenomic_histograms(
dat = echodata::BST1,
bigwig_metadata = echoannot::NOTT2019_bigwig_metadata[1:2,])
#> NOTT2019:: Creating epigenomic histograms plot
#> + Inferring genomic limits for window: 1x
#> Constructing GRanges query using min/max ranges across one or more chromosomes.
#> Downloading data from UCSC.
#> Importing... [1] exvivo_H3K27ac_tbp
#> Importing previously downloaded files: /github/home/.cache/R/echoannot/NOTT2019_epigenomic_peaks.rds
#> ++ NOTT2019:: 634,540 ranges retrieved.
#> dat is already a GRanges object.
#> 510 query SNP(s) detected with reference overlap.
#> + Calculating max histogram height
#> + Converting label units to Mb.
#> using coord:genome to parse x scale
#> using coord:genome to parse x scale
#> Warning: longer object length is not a multiple of shorter object length
#> Found more than one class "simpleUnit" in cache; using the first, from namespace 'hexbin'
#> Also defined by 'ggbio'
#> Found more than one class "unit" in cache; using the first, from namespace 'hexbin'
#> Also defined by 'ggbio'