Getting Started
¶ Author:
Brian M. Schilder ¶
¶ Updated:
Oct-30-2022 ¶
Source: vignettes/echoannot.Rmd
echoannot.Rmd## Registered S3 method overwritten by 'GGally':
## method from
## +.gg ggplot2Import data
To get the full dataset of all fine-mapped Parkinson’s Disease loci, you can use the following function:
merged_DT <- echodata::get_Nalls2019_merged()Annotate
Annotate SNP-wise fine-mapping results.
Here, we’re only annotating a small number of SNPs high-confidence
causal SNPs for demo purposes. The more SNPs you supply to
annotate_snps, the longer it will take to query the
selected databases for each SNP.
#### Only query high-confidence fine-mapping SNPs from one locus ####
dat <- merged_DT[Locus=="LRRK2" & Consensus_SNP==TRUE,]
#### Query annotations ####
dat_annot <- echoannot::annotate_snps(dat = dat,
haploreg_annotation = TRUE,
regulomeDB_annotation = TRUE,
biomart_annotation = TRUE) ## Loading required namespace: haploR## + Gathering annotation data from HaploReg...## ++ Submitting chunk 1 / 1## + Gathering annotation data from HaploReg...## ++ Submitting chunk 1 / 1## No encoding supplied: defaulting to UTF-8.
## No encoding supplied: defaulting to UTF-8.
## No encoding supplied: defaulting to UTF-8.
## No encoding supplied: defaulting to UTF-8.
## No encoding supplied: defaulting to UTF-8.## + Gathering annotation data from Biomart...
knitr::kable(dat_annot)| SNP | Locus | CHR | POS | P | Effect | StdErr | Freq | MAF | N_cases | N_controls | proportion_cases | A1 | A2 | leadSNP | ABF.CS | ABF.PP | SUSIE.CS | SUSIE.PP | POLYFUN_SUSIE.CS | POLYFUN_SUSIE.PP | FINEMAP.CS | FINEMAP.PP | Mb | Support | Consensus_SNP | mean.PP | mean.CS | N | t_stat | chr | pos_hg38 | r2 | D’ | is_query_snp | ref | alt | AFR | AMR | ASN | EUR | GERP_cons | SiPhy_cons | Chromatin_States | Chromatin_States_Imputed | Chromatin_Marks | DNAse | Proteins | eQTL | gwas | grasp | Motifs | GENCODE_id | GENCODE_name | GENCODE_direction | GENCODE_distance | RefSeq_id | RefSeq_name | RefSeq_direction | RefSeq_distance | dbSNP_functional_annotation | query_snp_rsid | Promoter_histone_marks | Enhancer_histone_marks | regulome.start | regulome.end | features.ChIP | features.DNase | features.Footprint | features.Footprint_matched | features.IC_matched_max | features.IC_max | features.PWM | features.PWM_matched | features.QTL | regulome_score.probability | regulome_score.ranking | allele | chr_name | chrom_start | chrom_end | chrom_strand | ensembl_gene_stable_id | consequence_type_tv | polyphen_prediction | polyphen_score | sift_prediction | sift_score | reg_consequence_types | validated |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs11175620 | LRRK2 | 12 | 40610864 | 0 | 0.1307 | 0.0142 | 0.1200 | 0.1200 | 56306 | 1417791 | 0.0382 | C | G | FALSE | NA | NA | 3 | 1 | 3 | 1 | NA | NA | 40.61086 | 2 | TRUE | 0.50 | 0 | 216621 | 9.204225 | 12 | 40217062 | 0 | 0 | 0 | G | C | 0.11 | 0.06 | 0.00 | 0.11 | 0 | 0 | E023,7_Enh;E024,7_Enh;E028,7_Enh;E029,7_Enh;E030,2_TssAFlnk;E031,2_TssAFlnk;E032,7_Enh;E035,1_TssA;E052,7_Enh;E080,7_Enh;E114,7_Enh;E117,7_Enh;E119,7_Enh;E124,2_TssAFlnk | E023,16_EnhW1;E025,19_DNase;E028,19_DNase;E029,14_EnhA2;E030,13_EnhA1;E031,14_EnhA2;E032,22_PromP;E033,16_EnhW1;E035,16_EnhW1;E036,14_EnhA2;E037,22_PromP;E038,22_PromP;E039,22_PromP;E040,22_PromP;E041,22_PromP;E042,22_PromP;E043,22_PromP;E044,22_PromP;E045,22_PromP;E046,22_PromP;E048,22_PromP;E050,22_PromP;E051,16_EnhW1;E057,22_PromP;E062,16_EnhW1;E071,22_PromP;E080,22_PromP;E086,19_DNase;E088,22_PromP;E112,17_EnhW2;E113,16_EnhW1;E114,19_DNase;E117,19_DNase;E124,13_EnhA1 | E011,H3K4me1_Enh;E018,H3K4me1_Enh;E023,H3K4me1_Enh;E024,H3K4me1_Enh;E025,H3K4me1_Enh;E028,H3K4me1_Enh;E029,H3K4me1_Enh;E030,H3K4me1_Enh;E031,H3K4me1_Enh;E032,H3K4me1_Enh;E036,H3K4me1_Enh;E049,H3K4me1_Enh;E050,H3K4me1_Enh;E052,H3K4me1_Enh;E056,H3K4me1_Enh;E057,H3K4me1_Enh;E080,H3K4me1_Enh;E086,H3K4me1_Enh;E088,H3K4me1_Enh;E113,H3K4me1_Enh;E114,H3K4me1_Enh;E117,H3K4me1_Enh;E119,H3K4me1_Enh;E124,H3K4me1_Enh;E023,H3K4me3_Pro;E030,H3K4me3_Pro;E031,H3K4me3_Pro;E035,H3K4me3_Pro;E109,H3K4me3_Pro;E124,H3K4me3_Pro;E025,H3K9ac_Pro;E029,H3K27ac_Enh;E032,H3K27ac_Enh;E080,H3K27ac_Enh;E113,H3K27ac_Enh;E124,H3K27ac_Enh | E006;E021;E029;E046;E080;E086;E088;E117;E120;E124 | . | GTEx2015_v6,Adipose_Subcutaneous,LRRK2,3.29211855956645e-07;GTEx2015_v6,Artery_Tibial,LRRK2,1.08781866153343e-11;GTEx2015_v6,Cells_Transformed_fibroblasts,LRRK2,1.72128347769633e-05;GTEx2015_v6,Nerve_Tibial,LRRK2,5.31856078341246e-10;GTEx2015_v6,Skin_Not_Sun_Exposed_Suprapubic,LRRK2,1.00796507632068e-05;GTEx2015_v6,Skin_Sun_Exposed_Lower_leg,LRRK2,1.06318553434241e-05;Westra2013,Whole_Blood,-,1.3808392059010189E-31;Westra2013,Whole_Blood,LRRK2,2.116806823753094E-5 | . | . | BCL_disc5;HDAC2_disc3 | ENSG00000225342.1 | AC079630.4 | 0 | 0 | NM_198578 | LRRK2 | 5 | 7947 | . | rs11175620 | BLD | FAT, ESC, BRST, BLD, MUS, ADRL, LNG, CRVX | 40610863 | 40610864 | TRUE | TRUE | FALSE | FALSE | 0 | 0 | FALSE | FALSE | FALSE | 0.60906 | 4 | G/C | 12 | 40610864 | 40610864 | 1 | ENSG00000225342 | non_coding_transcript_variant | NA | NA | NA | NA | 1000Genomes,Frequency,gnomAD,TOPMed | |
| rs11175620 | LRRK2 | 12 | 40610864 | 0 | 0.1307 | 0.0142 | 0.1200 | 0.1200 | 56306 | 1417791 | 0.0382 | C | G | FALSE | NA | NA | 3 | 1 | 3 | 1 | NA | NA | 40.61086 | 2 | TRUE | 0.50 | 0 | 216621 | 9.204225 | 12 | 40217062 | 0 | 0 | 0 | G | C | 0.11 | 0.06 | 0.00 | 0.11 | 0 | 0 | E023,7_Enh;E024,7_Enh;E028,7_Enh;E029,7_Enh;E030,2_TssAFlnk;E031,2_TssAFlnk;E032,7_Enh;E035,1_TssA;E052,7_Enh;E080,7_Enh;E114,7_Enh;E117,7_Enh;E119,7_Enh;E124,2_TssAFlnk | E023,16_EnhW1;E025,19_DNase;E028,19_DNase;E029,14_EnhA2;E030,13_EnhA1;E031,14_EnhA2;E032,22_PromP;E033,16_EnhW1;E035,16_EnhW1;E036,14_EnhA2;E037,22_PromP;E038,22_PromP;E039,22_PromP;E040,22_PromP;E041,22_PromP;E042,22_PromP;E043,22_PromP;E044,22_PromP;E045,22_PromP;E046,22_PromP;E048,22_PromP;E050,22_PromP;E051,16_EnhW1;E057,22_PromP;E062,16_EnhW1;E071,22_PromP;E080,22_PromP;E086,19_DNase;E088,22_PromP;E112,17_EnhW2;E113,16_EnhW1;E114,19_DNase;E117,19_DNase;E124,13_EnhA1 | E011,H3K4me1_Enh;E018,H3K4me1_Enh;E023,H3K4me1_Enh;E024,H3K4me1_Enh;E025,H3K4me1_Enh;E028,H3K4me1_Enh;E029,H3K4me1_Enh;E030,H3K4me1_Enh;E031,H3K4me1_Enh;E032,H3K4me1_Enh;E036,H3K4me1_Enh;E049,H3K4me1_Enh;E050,H3K4me1_Enh;E052,H3K4me1_Enh;E056,H3K4me1_Enh;E057,H3K4me1_Enh;E080,H3K4me1_Enh;E086,H3K4me1_Enh;E088,H3K4me1_Enh;E113,H3K4me1_Enh;E114,H3K4me1_Enh;E117,H3K4me1_Enh;E119,H3K4me1_Enh;E124,H3K4me1_Enh;E023,H3K4me3_Pro;E030,H3K4me3_Pro;E031,H3K4me3_Pro;E035,H3K4me3_Pro;E109,H3K4me3_Pro;E124,H3K4me3_Pro;E025,H3K9ac_Pro;E029,H3K27ac_Enh;E032,H3K27ac_Enh;E080,H3K27ac_Enh;E113,H3K27ac_Enh;E124,H3K27ac_Enh | E006;E021;E029;E046;E080;E086;E088;E117;E120;E124 | . | GTEx2015_v6,Adipose_Subcutaneous,LRRK2,3.29211855956645e-07;GTEx2015_v6,Artery_Tibial,LRRK2,1.08781866153343e-11;GTEx2015_v6,Cells_Transformed_fibroblasts,LRRK2,1.72128347769633e-05;GTEx2015_v6,Nerve_Tibial,LRRK2,5.31856078341246e-10;GTEx2015_v6,Skin_Not_Sun_Exposed_Suprapubic,LRRK2,1.00796507632068e-05;GTEx2015_v6,Skin_Sun_Exposed_Lower_leg,LRRK2,1.06318553434241e-05;Westra2013,Whole_Blood,-,1.3808392059010189E-31;Westra2013,Whole_Blood,LRRK2,2.116806823753094E-5 | . | . | BCL_disc5;HDAC2_disc3 | ENSG00000225342.1 | AC079630.4 | 0 | 0 | NM_198578 | LRRK2 | 5 | 7947 | . | rs11175620 | BLD | FAT, ESC, BRST, BLD, MUS, ADRL, LNG, CRVX | 40610863 | 40610864 | TRUE | TRUE | FALSE | FALSE | 0 | 0 | FALSE | FALSE | FALSE | 0.60906 | 4 | G/C | 12 | 40610864 | 40610864 | 1 | ENSG00000225342 | intron_variant | NA | NA | NA | NA | 1000Genomes,Frequency,gnomAD,TOPMed | |
| rs11175620 | LRRK2 | 12 | 40610864 | 0 | 0.1307 | 0.0142 | 0.1200 | 0.1200 | 56306 | 1417791 | 0.0382 | C | G | FALSE | NA | NA | 3 | 1 | 3 | 1 | NA | NA | 40.61086 | 2 | TRUE | 0.50 | 0 | 216621 | 9.204225 | 12 | 40217062 | 0 | 0 | 0 | G | C | 0.11 | 0.06 | 0.00 | 0.11 | 0 | 0 | E023,7_Enh;E024,7_Enh;E028,7_Enh;E029,7_Enh;E030,2_TssAFlnk;E031,2_TssAFlnk;E032,7_Enh;E035,1_TssA;E052,7_Enh;E080,7_Enh;E114,7_Enh;E117,7_Enh;E119,7_Enh;E124,2_TssAFlnk | E023,16_EnhW1;E025,19_DNase;E028,19_DNase;E029,14_EnhA2;E030,13_EnhA1;E031,14_EnhA2;E032,22_PromP;E033,16_EnhW1;E035,16_EnhW1;E036,14_EnhA2;E037,22_PromP;E038,22_PromP;E039,22_PromP;E040,22_PromP;E041,22_PromP;E042,22_PromP;E043,22_PromP;E044,22_PromP;E045,22_PromP;E046,22_PromP;E048,22_PromP;E050,22_PromP;E051,16_EnhW1;E057,22_PromP;E062,16_EnhW1;E071,22_PromP;E080,22_PromP;E086,19_DNase;E088,22_PromP;E112,17_EnhW2;E113,16_EnhW1;E114,19_DNase;E117,19_DNase;E124,13_EnhA1 | E011,H3K4me1_Enh;E018,H3K4me1_Enh;E023,H3K4me1_Enh;E024,H3K4me1_Enh;E025,H3K4me1_Enh;E028,H3K4me1_Enh;E029,H3K4me1_Enh;E030,H3K4me1_Enh;E031,H3K4me1_Enh;E032,H3K4me1_Enh;E036,H3K4me1_Enh;E049,H3K4me1_Enh;E050,H3K4me1_Enh;E052,H3K4me1_Enh;E056,H3K4me1_Enh;E057,H3K4me1_Enh;E080,H3K4me1_Enh;E086,H3K4me1_Enh;E088,H3K4me1_Enh;E113,H3K4me1_Enh;E114,H3K4me1_Enh;E117,H3K4me1_Enh;E119,H3K4me1_Enh;E124,H3K4me1_Enh;E023,H3K4me3_Pro;E030,H3K4me3_Pro;E031,H3K4me3_Pro;E035,H3K4me3_Pro;E109,H3K4me3_Pro;E124,H3K4me3_Pro;E025,H3K9ac_Pro;E029,H3K27ac_Enh;E032,H3K27ac_Enh;E080,H3K27ac_Enh;E113,H3K27ac_Enh;E124,H3K27ac_Enh | E006;E021;E029;E046;E080;E086;E088;E117;E120;E124 | . | GTEx2015_v6,Adipose_Subcutaneous,LRRK2,3.29211855956645e-07;GTEx2015_v6,Artery_Tibial,LRRK2,1.08781866153343e-11;GTEx2015_v6,Cells_Transformed_fibroblasts,LRRK2,1.72128347769633e-05;GTEx2015_v6,Nerve_Tibial,LRRK2,5.31856078341246e-10;GTEx2015_v6,Skin_Not_Sun_Exposed_Suprapubic,LRRK2,1.00796507632068e-05;GTEx2015_v6,Skin_Sun_Exposed_Lower_leg,LRRK2,1.06318553434241e-05;Westra2013,Whole_Blood,-,1.3808392059010189E-31;Westra2013,Whole_Blood,LRRK2,2.116806823753094E-5 | . | . | BCL_disc5;HDAC2_disc3 | ENSG00000225342.1 | AC079630.4 | 0 | 0 | NM_198578 | LRRK2 | 5 | 7947 | . | rs11175620 | BLD | FAT, ESC, BRST, BLD, MUS, ADRL, LNG, CRVX | 40610863 | 40610864 | TRUE | TRUE | FALSE | FALSE | 0 | 0 | FALSE | FALSE | FALSE | 0.60906 | 4 | G/C | 12 | 40610864 | 40610864 | 1 | ENSG00000188906 | intron_variant | NA | NA | NA | NA | 1000Genomes,Frequency,gnomAD,TOPMed | |
| rs7294619 | LRRK2 | 12 | 40617202 | 0 | -0.1276 | 0.0140 | 0.8783 | 0.1217 | 56306 | 1417791 | 0.0382 | T | C | FALSE | NA | NA | 2 | 1 | 2 | 1 | 1 | 1 | 40.61720 | 3 | TRUE | 0.75 | 0 | 216621 | -9.114286 | 12 | 40223400 | 0 | 0 | 0 | T | C | 0.11 | 0.06 | 0.00 | 0.11 | 0 | 0 | E023,7_Enh;E025,7_Enh;E029,1_TssA;E030,2_TssAFlnk;E031,2_TssAFlnk;E032,1_TssA;E033,1_TssA;E035,1_TssA;E046,1_TssA;E050,7_Enh;E051,1_TssA;E062,1_TssA;E063,7_Enh;E066,7_Enh;E068,7_Enh;E074,7_Enh;E076,7_Enh;E078,1_TssA;E086,1_TssA;E103,7_Enh;E116,10_TssBiv;E124,1_TssA | E009,23_PromBiv;E023,22_PromP;E025,22_PromP;E029,4_PromD2;E030,3_PromD1;E031,4_PromD2;E032,4_PromD2;E033,4_PromD2;E034,22_PromP;E035,4_PromD2;E036,4_PromD2;E037,22_PromP;E038,4_PromD2;E039,12_TxEnhW;E040,12_TxEnhW;E041,12_TxEnhW;E042,22_PromP;E043,4_PromD2;E044,4_PromD2;E045,12_TxEnhW;E046,4_PromD2;E047,4_PromD2;E048,22_PromP;E050,4_PromD2;E051,4_PromD2;E052,19_DNase;E053,22_PromP;E055,22_PromP;E057,22_PromP;E061,22_PromP;E062,4_PromD2;E063,22_PromP;E066,4_PromD2;E067,22_PromP;E068,22_PromP;E069,22_PromP;E070,22_PromP;E071,12_TxEnhW;E072,22_PromP;E073,22_PromP;E074,19_DNase;E075,22_PromP;E076,4_PromD2;E077,4_PromD2;E078,4_PromD2;E080,22_PromP;E081,22_PromP;E084,22_PromP;E085,22_PromP;E086,22_PromP;E088,22_PromP;E091,22_PromP;E092,22_PromP;E093,22_PromP;E100,22_PromP;E101,4_PromD2;E102,4_PromD2;E103,22_PromP;E106,22_PromP;E108,22_PromP;E109,22_PromP;E110,22_PromP;E111,16_EnhW1;E112,4_PromD2;E113,4_PromD2;E114,22_PromP;E115,23_PromBiv;E116,4_PromD2;E118,22_PromP;E119,22_PromP;E122,22_PromP;E124,3_PromD1 | E023,H3K4me1_Enh;E025,H3K4me1_Enh;E026,H3K4me1_Enh;E027,H3K4me1_Enh;E028,H3K4me1_Enh;E030,H3K4me1_Enh;E031,H3K4me1_Enh;E032,H3K4me1_Enh;E035,H3K4me1_Enh;E036,H3K4me1_Enh;E038,H3K4me1_Enh;E041,H3K4me1_Enh;E043,H3K4me1_Enh;E047,H3K4me1_Enh;E048,H3K4me1_Enh;E049,H3K4me1_Enh;E050,H3K4me1_Enh;E051,H3K4me1_Enh;E052,H3K4me1_Enh;E061,H3K4me1_Enh;E062,H3K4me1_Enh;E063,H3K4me1_Enh;E066,H3K4me1_Enh;E067,H3K4me1_Enh;E068,H3K4me1_Enh;E069,H3K4me1_Enh;E071,H3K4me1_Enh;E072,H3K4me1_Enh;E073,H3K4me1_Enh;E074,H3K4me1_Enh;E076,H3K4me1_Enh;E077,H3K4me1_Enh;E078,H3K4me1_Enh;E080,H3K4me1_Enh;E087,H3K4me1_Enh;E102,H3K4me1_Enh;E103,H3K4me1_Enh;E108,H3K4me1_Enh;E109,H3K4me1_Enh;E111,H3K4me1_Enh;E114,H3K4me1_Enh;E124,H3K4me1_Enh;E129,H3K4me1_Enh;E023,H3K4me3_Pro;E024,H3K4me3_Pro;E025,H3K4me3_Pro;E029,H3K4me3_Pro;E030,H3K4me3_Pro;E031,H3K4me3_Pro;E032,H3K4me3_Pro;E033,H3K4me3_Pro;E035,H3K4me3_Pro;E036,H3K4me3_Pro;E039,H3K4me3_Pro;E041,H3K4me3_Pro;E043,H3K4me3_Pro;E045,H3K4me3_Pro;E046,H3K4me3_Pro;E047,H3K4me3_Pro;E049,H3K4me3_Pro;E051,H3K4me3_Pro;E052,H3K4me3_Pro;E062,H3K4me3_Pro;E063,H3K4me3_Pro;E065,H3K4me3_Pro;E066,H3K4me3_Pro;E067,H3K4me3_Pro;E068,H3K4me3_Pro;E071,H3K4me3_Pro;E072,H3K4me3_Pro;E073,H3K4me3_Pro;E074,H3K4me3_Pro;E076,H3K4me3_Pro;E077,H3K4me3_Pro;E078,H3K4me3_Pro;E084,H3K4me3_Pro;E086,H3K4me3_Pro;E088,H3K4me3_Pro;E091,H3K4me3_Pro;E093,H3K4me3_Pro;E101,H3K4me3_Pro;E102,H3K4me3_Pro;E103,H3K4me3_Pro;E104,H3K4me3_Pro;E105,H3K4me3_Pro;E108,H3K4me3_Pro;E111,H3K4me3_Pro;E112,H3K4me3_Pro;E114,H3K4me3_Pro;E116,H3K4me3_Pro;E124,H3K4me3_Pro;E126,H3K4me3_Pro;E023,H3K9ac_Pro;E025,H3K9ac_Pro;E026,H3K9ac_Pro;E052,H3K9ac_Pro;E062,H3K9ac_Pro;E063,H3K9ac_Pro;E066,H3K9ac_Pro;E068,H3K9ac_Pro;E076,H3K9ac_Pro;E077,H3K9ac_Pro;E086,H3K9ac_Pro;E101,H3K9ac_Pro;E102,H3K9ac_Pro;E116,H3K9ac_Pro;E124,H3K9ac_Pro;E029,H3K27ac_Enh;E032,H3K27ac_Enh;E046,H3K27ac_Enh;E062,H3K27ac_Enh;E063,H3K27ac_Enh;E066,H3K27ac_Enh;E067,H3K27ac_Enh;E068,H3K27ac_Enh;E069,H3K27ac_Enh;E072,H3K27ac_Enh;E073,H3K27ac_Enh;E074,H3K27ac_Enh;E075,H3K27ac_Enh;E076,H3K27ac_Enh;E078,H3K27ac_Enh;E084,H3K27ac_Enh;E093,H3K27ac_Enh;E101,H3K27ac_Enh;E102,H3K27ac_Enh;E103,H3K27ac_Enh;E116,H3K27ac_Enh;E124,H3K27ac_Enh;E126,H3K27ac_Enh | . | GTEx2015_v6,Adipose_Subcutaneous,LRRK2,4.21086496303076e-07;GTEx2015_v6,Artery_Tibial,LRRK2,7.12887180198536e-12;GTEx2015_v6,Cells_Transformed_fibroblasts,LRRK2,1.24729820163706e-05;GTEx2015_v6,Nerve_Tibial,LRRK2,5.97959499940652e-10;GTEx2015_v6,Skin_Not_Sun_Exposed_Suprapubic,LRRK2,1.00796507632068e-05;GTEx2015_v6,Skin_Sun_Exposed_Lower_leg,LRRK2,1.08361107066017e-05;Westra2013,Whole_Blood,-,1.9330946764156724E-31;Westra2013,Whole_Blood,LRRK2,1.807765948958669E-5 | . | . | Barx1;Barx2;CDP_2;En-1_1;Hoxb6;Irf_known7;Ncx_1;Pou5f1_known2;Sox_18;Sox_3;Sox_9 | ENSG00000225342.1 | AC079630.4 | 0 | 0 | NM_198578 | LRRK2 | 5 | 1609 | . | rs7294619 | BLD, GI, KID | FAT, BLD, LIV, BRN, GI | 40617201 | 40617202 | FALSE | TRUE | FALSE | FALSE | 0 | 0.939999997615814 | TRUE | FALSE | FALSE | 0.07617 | 5 | T/C | 12 | 40617202 | 40617202 | 1 | ENSG00000225342 | non_coding_transcript_variant | NA | NA | NA | NA | regulatory_region_variant | 1000Genomes,Cited,Frequency,gnomAD,TOPMed | |
| rs7294619 | LRRK2 | 12 | 40617202 | 0 | -0.1276 | 0.0140 | 0.8783 | 0.1217 | 56306 | 1417791 | 0.0382 | T | C | FALSE | NA | NA | 2 | 1 | 2 | 1 | 1 | 1 | 40.61720 | 3 | TRUE | 0.75 | 0 | 216621 | -9.114286 | 12 | 40223400 | 0 | 0 | 0 | T | C | 0.11 | 0.06 | 0.00 | 0.11 | 0 | 0 | E023,7_Enh;E025,7_Enh;E029,1_TssA;E030,2_TssAFlnk;E031,2_TssAFlnk;E032,1_TssA;E033,1_TssA;E035,1_TssA;E046,1_TssA;E050,7_Enh;E051,1_TssA;E062,1_TssA;E063,7_Enh;E066,7_Enh;E068,7_Enh;E074,7_Enh;E076,7_Enh;E078,1_TssA;E086,1_TssA;E103,7_Enh;E116,10_TssBiv;E124,1_TssA | E009,23_PromBiv;E023,22_PromP;E025,22_PromP;E029,4_PromD2;E030,3_PromD1;E031,4_PromD2;E032,4_PromD2;E033,4_PromD2;E034,22_PromP;E035,4_PromD2;E036,4_PromD2;E037,22_PromP;E038,4_PromD2;E039,12_TxEnhW;E040,12_TxEnhW;E041,12_TxEnhW;E042,22_PromP;E043,4_PromD2;E044,4_PromD2;E045,12_TxEnhW;E046,4_PromD2;E047,4_PromD2;E048,22_PromP;E050,4_PromD2;E051,4_PromD2;E052,19_DNase;E053,22_PromP;E055,22_PromP;E057,22_PromP;E061,22_PromP;E062,4_PromD2;E063,22_PromP;E066,4_PromD2;E067,22_PromP;E068,22_PromP;E069,22_PromP;E070,22_PromP;E071,12_TxEnhW;E072,22_PromP;E073,22_PromP;E074,19_DNase;E075,22_PromP;E076,4_PromD2;E077,4_PromD2;E078,4_PromD2;E080,22_PromP;E081,22_PromP;E084,22_PromP;E085,22_PromP;E086,22_PromP;E088,22_PromP;E091,22_PromP;E092,22_PromP;E093,22_PromP;E100,22_PromP;E101,4_PromD2;E102,4_PromD2;E103,22_PromP;E106,22_PromP;E108,22_PromP;E109,22_PromP;E110,22_PromP;E111,16_EnhW1;E112,4_PromD2;E113,4_PromD2;E114,22_PromP;E115,23_PromBiv;E116,4_PromD2;E118,22_PromP;E119,22_PromP;E122,22_PromP;E124,3_PromD1 | E023,H3K4me1_Enh;E025,H3K4me1_Enh;E026,H3K4me1_Enh;E027,H3K4me1_Enh;E028,H3K4me1_Enh;E030,H3K4me1_Enh;E031,H3K4me1_Enh;E032,H3K4me1_Enh;E035,H3K4me1_Enh;E036,H3K4me1_Enh;E038,H3K4me1_Enh;E041,H3K4me1_Enh;E043,H3K4me1_Enh;E047,H3K4me1_Enh;E048,H3K4me1_Enh;E049,H3K4me1_Enh;E050,H3K4me1_Enh;E051,H3K4me1_Enh;E052,H3K4me1_Enh;E061,H3K4me1_Enh;E062,H3K4me1_Enh;E063,H3K4me1_Enh;E066,H3K4me1_Enh;E067,H3K4me1_Enh;E068,H3K4me1_Enh;E069,H3K4me1_Enh;E071,H3K4me1_Enh;E072,H3K4me1_Enh;E073,H3K4me1_Enh;E074,H3K4me1_Enh;E076,H3K4me1_Enh;E077,H3K4me1_Enh;E078,H3K4me1_Enh;E080,H3K4me1_Enh;E087,H3K4me1_Enh;E102,H3K4me1_Enh;E103,H3K4me1_Enh;E108,H3K4me1_Enh;E109,H3K4me1_Enh;E111,H3K4me1_Enh;E114,H3K4me1_Enh;E124,H3K4me1_Enh;E129,H3K4me1_Enh;E023,H3K4me3_Pro;E024,H3K4me3_Pro;E025,H3K4me3_Pro;E029,H3K4me3_Pro;E030,H3K4me3_Pro;E031,H3K4me3_Pro;E032,H3K4me3_Pro;E033,H3K4me3_Pro;E035,H3K4me3_Pro;E036,H3K4me3_Pro;E039,H3K4me3_Pro;E041,H3K4me3_Pro;E043,H3K4me3_Pro;E045,H3K4me3_Pro;E046,H3K4me3_Pro;E047,H3K4me3_Pro;E049,H3K4me3_Pro;E051,H3K4me3_Pro;E052,H3K4me3_Pro;E062,H3K4me3_Pro;E063,H3K4me3_Pro;E065,H3K4me3_Pro;E066,H3K4me3_Pro;E067,H3K4me3_Pro;E068,H3K4me3_Pro;E071,H3K4me3_Pro;E072,H3K4me3_Pro;E073,H3K4me3_Pro;E074,H3K4me3_Pro;E076,H3K4me3_Pro;E077,H3K4me3_Pro;E078,H3K4me3_Pro;E084,H3K4me3_Pro;E086,H3K4me3_Pro;E088,H3K4me3_Pro;E091,H3K4me3_Pro;E093,H3K4me3_Pro;E101,H3K4me3_Pro;E102,H3K4me3_Pro;E103,H3K4me3_Pro;E104,H3K4me3_Pro;E105,H3K4me3_Pro;E108,H3K4me3_Pro;E111,H3K4me3_Pro;E112,H3K4me3_Pro;E114,H3K4me3_Pro;E116,H3K4me3_Pro;E124,H3K4me3_Pro;E126,H3K4me3_Pro;E023,H3K9ac_Pro;E025,H3K9ac_Pro;E026,H3K9ac_Pro;E052,H3K9ac_Pro;E062,H3K9ac_Pro;E063,H3K9ac_Pro;E066,H3K9ac_Pro;E068,H3K9ac_Pro;E076,H3K9ac_Pro;E077,H3K9ac_Pro;E086,H3K9ac_Pro;E101,H3K9ac_Pro;E102,H3K9ac_Pro;E116,H3K9ac_Pro;E124,H3K9ac_Pro;E029,H3K27ac_Enh;E032,H3K27ac_Enh;E046,H3K27ac_Enh;E062,H3K27ac_Enh;E063,H3K27ac_Enh;E066,H3K27ac_Enh;E067,H3K27ac_Enh;E068,H3K27ac_Enh;E069,H3K27ac_Enh;E072,H3K27ac_Enh;E073,H3K27ac_Enh;E074,H3K27ac_Enh;E075,H3K27ac_Enh;E076,H3K27ac_Enh;E078,H3K27ac_Enh;E084,H3K27ac_Enh;E093,H3K27ac_Enh;E101,H3K27ac_Enh;E102,H3K27ac_Enh;E103,H3K27ac_Enh;E116,H3K27ac_Enh;E124,H3K27ac_Enh;E126,H3K27ac_Enh | . | GTEx2015_v6,Adipose_Subcutaneous,LRRK2,4.21086496303076e-07;GTEx2015_v6,Artery_Tibial,LRRK2,7.12887180198536e-12;GTEx2015_v6,Cells_Transformed_fibroblasts,LRRK2,1.24729820163706e-05;GTEx2015_v6,Nerve_Tibial,LRRK2,5.97959499940652e-10;GTEx2015_v6,Skin_Not_Sun_Exposed_Suprapubic,LRRK2,1.00796507632068e-05;GTEx2015_v6,Skin_Sun_Exposed_Lower_leg,LRRK2,1.08361107066017e-05;Westra2013,Whole_Blood,-,1.9330946764156724E-31;Westra2013,Whole_Blood,LRRK2,1.807765948958669E-5 | . | . | Barx1;Barx2;CDP_2;En-1_1;Hoxb6;Irf_known7;Ncx_1;Pou5f1_known2;Sox_18;Sox_3;Sox_9 | ENSG00000225342.1 | AC079630.4 | 0 | 0 | NM_198578 | LRRK2 | 5 | 1609 | . | rs7294619 | BLD, GI, KID | FAT, BLD, LIV, BRN, GI | 40617201 | 40617202 | FALSE | TRUE | FALSE | FALSE | 0 | 0.939999997615814 | TRUE | FALSE | FALSE | 0.07617 | 5 | T/C | 12 | 40617202 | 40617202 | 1 | ENSG00000225342 | intron_variant | NA | NA | NA | NA | regulatory_region_variant | 1000Genomes,Cited,Frequency,gnomAD,TOPMed | |
| rs7294619 | LRRK2 | 12 | 40617202 | 0 | -0.1276 | 0.0140 | 0.8783 | 0.1217 | 56306 | 1417791 | 0.0382 | T | C | FALSE | NA | NA | 2 | 1 | 2 | 1 | 1 | 1 | 40.61720 | 3 | TRUE | 0.75 | 0 | 216621 | -9.114286 | 12 | 40223400 | 0 | 0 | 0 | T | C | 0.11 | 0.06 | 0.00 | 0.11 | 0 | 0 | E023,7_Enh;E025,7_Enh;E029,1_TssA;E030,2_TssAFlnk;E031,2_TssAFlnk;E032,1_TssA;E033,1_TssA;E035,1_TssA;E046,1_TssA;E050,7_Enh;E051,1_TssA;E062,1_TssA;E063,7_Enh;E066,7_Enh;E068,7_Enh;E074,7_Enh;E076,7_Enh;E078,1_TssA;E086,1_TssA;E103,7_Enh;E116,10_TssBiv;E124,1_TssA | E009,23_PromBiv;E023,22_PromP;E025,22_PromP;E029,4_PromD2;E030,3_PromD1;E031,4_PromD2;E032,4_PromD2;E033,4_PromD2;E034,22_PromP;E035,4_PromD2;E036,4_PromD2;E037,22_PromP;E038,4_PromD2;E039,12_TxEnhW;E040,12_TxEnhW;E041,12_TxEnhW;E042,22_PromP;E043,4_PromD2;E044,4_PromD2;E045,12_TxEnhW;E046,4_PromD2;E047,4_PromD2;E048,22_PromP;E050,4_PromD2;E051,4_PromD2;E052,19_DNase;E053,22_PromP;E055,22_PromP;E057,22_PromP;E061,22_PromP;E062,4_PromD2;E063,22_PromP;E066,4_PromD2;E067,22_PromP;E068,22_PromP;E069,22_PromP;E070,22_PromP;E071,12_TxEnhW;E072,22_PromP;E073,22_PromP;E074,19_DNase;E075,22_PromP;E076,4_PromD2;E077,4_PromD2;E078,4_PromD2;E080,22_PromP;E081,22_PromP;E084,22_PromP;E085,22_PromP;E086,22_PromP;E088,22_PromP;E091,22_PromP;E092,22_PromP;E093,22_PromP;E100,22_PromP;E101,4_PromD2;E102,4_PromD2;E103,22_PromP;E106,22_PromP;E108,22_PromP;E109,22_PromP;E110,22_PromP;E111,16_EnhW1;E112,4_PromD2;E113,4_PromD2;E114,22_PromP;E115,23_PromBiv;E116,4_PromD2;E118,22_PromP;E119,22_PromP;E122,22_PromP;E124,3_PromD1 | E023,H3K4me1_Enh;E025,H3K4me1_Enh;E026,H3K4me1_Enh;E027,H3K4me1_Enh;E028,H3K4me1_Enh;E030,H3K4me1_Enh;E031,H3K4me1_Enh;E032,H3K4me1_Enh;E035,H3K4me1_Enh;E036,H3K4me1_Enh;E038,H3K4me1_Enh;E041,H3K4me1_Enh;E043,H3K4me1_Enh;E047,H3K4me1_Enh;E048,H3K4me1_Enh;E049,H3K4me1_Enh;E050,H3K4me1_Enh;E051,H3K4me1_Enh;E052,H3K4me1_Enh;E061,H3K4me1_Enh;E062,H3K4me1_Enh;E063,H3K4me1_Enh;E066,H3K4me1_Enh;E067,H3K4me1_Enh;E068,H3K4me1_Enh;E069,H3K4me1_Enh;E071,H3K4me1_Enh;E072,H3K4me1_Enh;E073,H3K4me1_Enh;E074,H3K4me1_Enh;E076,H3K4me1_Enh;E077,H3K4me1_Enh;E078,H3K4me1_Enh;E080,H3K4me1_Enh;E087,H3K4me1_Enh;E102,H3K4me1_Enh;E103,H3K4me1_Enh;E108,H3K4me1_Enh;E109,H3K4me1_Enh;E111,H3K4me1_Enh;E114,H3K4me1_Enh;E124,H3K4me1_Enh;E129,H3K4me1_Enh;E023,H3K4me3_Pro;E024,H3K4me3_Pro;E025,H3K4me3_Pro;E029,H3K4me3_Pro;E030,H3K4me3_Pro;E031,H3K4me3_Pro;E032,H3K4me3_Pro;E033,H3K4me3_Pro;E035,H3K4me3_Pro;E036,H3K4me3_Pro;E039,H3K4me3_Pro;E041,H3K4me3_Pro;E043,H3K4me3_Pro;E045,H3K4me3_Pro;E046,H3K4me3_Pro;E047,H3K4me3_Pro;E049,H3K4me3_Pro;E051,H3K4me3_Pro;E052,H3K4me3_Pro;E062,H3K4me3_Pro;E063,H3K4me3_Pro;E065,H3K4me3_Pro;E066,H3K4me3_Pro;E067,H3K4me3_Pro;E068,H3K4me3_Pro;E071,H3K4me3_Pro;E072,H3K4me3_Pro;E073,H3K4me3_Pro;E074,H3K4me3_Pro;E076,H3K4me3_Pro;E077,H3K4me3_Pro;E078,H3K4me3_Pro;E084,H3K4me3_Pro;E086,H3K4me3_Pro;E088,H3K4me3_Pro;E091,H3K4me3_Pro;E093,H3K4me3_Pro;E101,H3K4me3_Pro;E102,H3K4me3_Pro;E103,H3K4me3_Pro;E104,H3K4me3_Pro;E105,H3K4me3_Pro;E108,H3K4me3_Pro;E111,H3K4me3_Pro;E112,H3K4me3_Pro;E114,H3K4me3_Pro;E116,H3K4me3_Pro;E124,H3K4me3_Pro;E126,H3K4me3_Pro;E023,H3K9ac_Pro;E025,H3K9ac_Pro;E026,H3K9ac_Pro;E052,H3K9ac_Pro;E062,H3K9ac_Pro;E063,H3K9ac_Pro;E066,H3K9ac_Pro;E068,H3K9ac_Pro;E076,H3K9ac_Pro;E077,H3K9ac_Pro;E086,H3K9ac_Pro;E101,H3K9ac_Pro;E102,H3K9ac_Pro;E116,H3K9ac_Pro;E124,H3K9ac_Pro;E029,H3K27ac_Enh;E032,H3K27ac_Enh;E046,H3K27ac_Enh;E062,H3K27ac_Enh;E063,H3K27ac_Enh;E066,H3K27ac_Enh;E067,H3K27ac_Enh;E068,H3K27ac_Enh;E069,H3K27ac_Enh;E072,H3K27ac_Enh;E073,H3K27ac_Enh;E074,H3K27ac_Enh;E075,H3K27ac_Enh;E076,H3K27ac_Enh;E078,H3K27ac_Enh;E084,H3K27ac_Enh;E093,H3K27ac_Enh;E101,H3K27ac_Enh;E102,H3K27ac_Enh;E103,H3K27ac_Enh;E116,H3K27ac_Enh;E124,H3K27ac_Enh;E126,H3K27ac_Enh | . | GTEx2015_v6,Adipose_Subcutaneous,LRRK2,4.21086496303076e-07;GTEx2015_v6,Artery_Tibial,LRRK2,7.12887180198536e-12;GTEx2015_v6,Cells_Transformed_fibroblasts,LRRK2,1.24729820163706e-05;GTEx2015_v6,Nerve_Tibial,LRRK2,5.97959499940652e-10;GTEx2015_v6,Skin_Not_Sun_Exposed_Suprapubic,LRRK2,1.00796507632068e-05;GTEx2015_v6,Skin_Sun_Exposed_Lower_leg,LRRK2,1.08361107066017e-05;Westra2013,Whole_Blood,-,1.9330946764156724E-31;Westra2013,Whole_Blood,LRRK2,1.807765948958669E-5 | . | . | Barx1;Barx2;CDP_2;En-1_1;Hoxb6;Irf_known7;Ncx_1;Pou5f1_known2;Sox_18;Sox_3;Sox_9 | ENSG00000225342.1 | AC079630.4 | 0 | 0 | NM_198578 | LRRK2 | 5 | 1609 | . | rs7294619 | BLD, GI, KID | FAT, BLD, LIV, BRN, GI | 40617201 | 40617202 | FALSE | TRUE | FALSE | FALSE | 0 | 0.939999997615814 | TRUE | FALSE | FALSE | 0.07617 | 5 | T/C | 12 | 40617202 | 40617202 | 1 | ENSG00000188906 | intron_variant | NA | NA | NA | NA | regulatory_region_variant | 1000Genomes,Cited,Frequency,gnomAD,TOPMed | |
| rs74324737 | LRRK2 | 12 | 40625081 | 0 | 0.1246 | 0.0141 | 0.1216 | 0.1216 | 56306 | 1417791 | 0.0382 | A | G | FALSE | NA | NA | 4 | 1 | 4 | 1 | NA | NA | 40.62508 | 2 | TRUE | 0.50 | 0 | 216621 | 8.836879 | 12 | 40231279 | 0 | 0 | 0 | G | A | 0.09 | 0.06 | 0.00 | 0.11 | 0 | 0 | E029,6_EnhG;E030,6_EnhG;E031,7_Enh;E032,6_EnhG;E080,7_Enh;E124,6_EnhG | E029,10_TxEnh5;E030,10_TxEnh5;E031,10_TxEnh5;E032,10_TxEnh5;E124,10_TxEnh5 | E029,H3K27ac_Enh;E032,H3K27ac_Enh;E046,H3K27ac_Enh;E068,H3K27ac_Enh;E080,H3K27ac_Enh;E124,H3K27ac_Enh;E029,H3K4me1_Enh;E030,H3K4me1_Enh;E031,H3K4me1_Enh;E032,H3K4me1_Enh;E067,H3K4me1_Enh;E073,H3K4me1_Enh;E078,H3K4me1_Enh;E116,H3K4me1_Enh;E124,H3K4me1_Enh;E031,H3K4me3_Pro;E067,H3K4me3_Pro;E116,H3K4me3_Pro;E124,H3K4me3_Pro;E124,H3K9ac_Pro | . | GTEx2015_v6,Adipose_Subcutaneous,LRRK2,7.71727072375557e-07;GTEx2015_v6,Artery_Tibial,LRRK2,3.79436480774191e-12;GTEx2015_v6,Cells_Transformed_fibroblasts,LRRK2,6.94075809560474e-06;GTEx2015_v6,Nerve_Tibial,LRRK2,8.74398504351068e-10;GTEx2015_v6,Skin_Not_Sun_Exposed_Suprapubic,LRRK2,9.31129758632871e-06;GTEx2015_v6,Skin_Sun_Exposed_Lower_leg,LRRK2,1.18501232307152e-05 | . | . | E2A_3;E2A_5;Lmo2-complex_1;Mtf1_1;Myf_1;Myf_3;NF-E2_disc3;TCF12_disc1;ZEB1_known3 | ENSG00000188906.9 | LRRK2 | 0 | 0 | NM_198578 | LRRK2 | 0 | 0 | INT | rs74324737 | BLD, ADRL | 40625080 | 40625081 | FALSE | TRUE | FALSE | FALSE | 0 | 1.79999995231628 | TRUE | FALSE | FALSE | 0.9223 | 5 | G/A | 12 | 40625081 | 40625081 | 1 | ENSG00000188906 | intron_variant | NA | NA | NA | NA | 1000Genomes,Cited,Frequency,gnomAD,TOPMed | |||
| rs74324737 | LRRK2 | 12 | 40625081 | 0 | 0.1246 | 0.0141 | 0.1216 | 0.1216 | 56306 | 1417791 | 0.0382 | A | G | FALSE | NA | NA | 4 | 1 | 4 | 1 | NA | NA | 40.62508 | 2 | TRUE | 0.50 | 0 | 216621 | 8.836879 | 12 | 40231279 | 0 | 0 | 0 | G | A | 0.09 | 0.06 | 0.00 | 0.11 | 0 | 0 | E029,6_EnhG;E030,6_EnhG;E031,7_Enh;E032,6_EnhG;E080,7_Enh;E124,6_EnhG | E029,10_TxEnh5;E030,10_TxEnh5;E031,10_TxEnh5;E032,10_TxEnh5;E124,10_TxEnh5 | E029,H3K27ac_Enh;E032,H3K27ac_Enh;E046,H3K27ac_Enh;E068,H3K27ac_Enh;E080,H3K27ac_Enh;E124,H3K27ac_Enh;E029,H3K4me1_Enh;E030,H3K4me1_Enh;E031,H3K4me1_Enh;E032,H3K4me1_Enh;E067,H3K4me1_Enh;E073,H3K4me1_Enh;E078,H3K4me1_Enh;E116,H3K4me1_Enh;E124,H3K4me1_Enh;E031,H3K4me3_Pro;E067,H3K4me3_Pro;E116,H3K4me3_Pro;E124,H3K4me3_Pro;E124,H3K9ac_Pro | . | GTEx2015_v6,Adipose_Subcutaneous,LRRK2,7.71727072375557e-07;GTEx2015_v6,Artery_Tibial,LRRK2,3.79436480774191e-12;GTEx2015_v6,Cells_Transformed_fibroblasts,LRRK2,6.94075809560474e-06;GTEx2015_v6,Nerve_Tibial,LRRK2,8.74398504351068e-10;GTEx2015_v6,Skin_Not_Sun_Exposed_Suprapubic,LRRK2,9.31129758632871e-06;GTEx2015_v6,Skin_Sun_Exposed_Lower_leg,LRRK2,1.18501232307152e-05 | . | . | E2A_3;E2A_5;Lmo2-complex_1;Mtf1_1;Myf_1;Myf_3;NF-E2_disc3;TCF12_disc1;ZEB1_known3 | ENSG00000188906.9 | LRRK2 | 0 | 0 | NM_198578 | LRRK2 | 0 | 0 | INT | rs74324737 | BLD, ADRL | 40625080 | 40625081 | FALSE | TRUE | FALSE | FALSE | 0 | 1.79999995231628 | TRUE | FALSE | FALSE | 0.9223 | 5 | G/A | 12 | 40625081 | 40625081 | 1 | ENSG00000188906 | non_coding_transcript_variant | NA | NA | NA | NA | 1000Genomes,Cited,Frequency,gnomAD,TOPMed | |||
| rs76904798 | LRRK2 | 12 | 40614434 | 0 | 0.1439 | 0.0130 | 0.1444 | 0.1444 | 56306 | 1417791 | 0.0382 | T | C | TRUE | NA | NA | 1 | 1 | 1 | 1 | 1 | 1 | 40.61443 | 3 | TRUE | 0.75 | 0 | 216621 | 11.069231 | 12 | 40220632 | 0 | 0 | 0 | C | T | 0.11 | 0.21 | 0.03 | 0.13 | 0 | 0 | E029,7_Enh;E030,7_Enh;E124,7_Enh | E029,H3K4me1_Enh;E030,H3K4me1_Enh;E031,H3K4me1_Enh;E124,H3K4me1_Enh;E124,H3K27ac_Enh | . | GTEx2015_v6,Adipose_Subcutaneous,LRRK2,2.6136557196757e-09;GTEx2015_v6,Artery_Coronary,LRRK2,8.10828434519411e-08;GTEx2015_v6,Artery_Tibial,LRRK2,1.29589062525083e-14;GTEx2015_v6,Cells_Transformed_fibroblasts,LRRK2,2.01546552400868e-05;GTEx2015_v6,Esophagus_Muscularis,LRRK2,1.17034509288903e-05;GTEx2015_v6,Nerve_Tibial,LRRK2,1.17833931025132e-15;GTEx2015_v6,Skin_Not_Sun_Exposed_Suprapubic,LRRK2,3.02678179339592e-06;GTEx2015_v6,Skin_Sun_Exposed_Lower_leg,LRRK2,1.21658440247309e-07 | 25064009,Parkinson’s disease,5E-14 | . | Dbx1;Hmx_2;Hoxb8;Hoxd8;Ncx_2;Nkx6-1_3;Pou2f2_known2;Sox_14;Sox_15;Sox_16;Sox_18;Sox_19;Sox_2;Sox_5;Sox_7;Sox_9 | ENSG00000225342.1 | AC079630.4 | 0 | 0 | NM_198578 | LRRK2 | 5 | 4377 | . | rs76904798 | BLD | 40614433 | 40614434 | FALSE | FALSE | FALSE | FALSE | 0 | 0.230000004172325 | TRUE | FALSE | FALSE | 0.14464 | 6 | C/T | 12 | 40614434 | 40614434 | 1 | ENSG00000225342 | non_coding_transcript_variant | NA | NA | NA | NA | 1000Genomes,Cited,Frequency,gnomAD,Phenotype_or_Disease,TOPMed | ||||
| rs76904798 | LRRK2 | 12 | 40614434 | 0 | 0.1439 | 0.0130 | 0.1444 | 0.1444 | 56306 | 1417791 | 0.0382 | T | C | TRUE | NA | NA | 1 | 1 | 1 | 1 | 1 | 1 | 40.61443 | 3 | TRUE | 0.75 | 0 | 216621 | 11.069231 | 12 | 40220632 | 0 | 0 | 0 | C | T | 0.11 | 0.21 | 0.03 | 0.13 | 0 | 0 | E029,7_Enh;E030,7_Enh;E124,7_Enh | E029,H3K4me1_Enh;E030,H3K4me1_Enh;E031,H3K4me1_Enh;E124,H3K4me1_Enh;E124,H3K27ac_Enh | . | GTEx2015_v6,Adipose_Subcutaneous,LRRK2,2.6136557196757e-09;GTEx2015_v6,Artery_Coronary,LRRK2,8.10828434519411e-08;GTEx2015_v6,Artery_Tibial,LRRK2,1.29589062525083e-14;GTEx2015_v6,Cells_Transformed_fibroblasts,LRRK2,2.01546552400868e-05;GTEx2015_v6,Esophagus_Muscularis,LRRK2,1.17034509288903e-05;GTEx2015_v6,Nerve_Tibial,LRRK2,1.17833931025132e-15;GTEx2015_v6,Skin_Not_Sun_Exposed_Suprapubic,LRRK2,3.02678179339592e-06;GTEx2015_v6,Skin_Sun_Exposed_Lower_leg,LRRK2,1.21658440247309e-07 | 25064009,Parkinson’s disease,5E-14 | . | Dbx1;Hmx_2;Hoxb8;Hoxd8;Ncx_2;Nkx6-1_3;Pou2f2_known2;Sox_14;Sox_15;Sox_16;Sox_18;Sox_19;Sox_2;Sox_5;Sox_7;Sox_9 | ENSG00000225342.1 | AC079630.4 | 0 | 0 | NM_198578 | LRRK2 | 5 | 4377 | . | rs76904798 | BLD | 40614433 | 40614434 | FALSE | FALSE | FALSE | FALSE | 0 | 0.230000004172325 | TRUE | FALSE | FALSE | 0.14464 | 6 | C/T | 12 | 40614434 | 40614434 | 1 | ENSG00000225342 | intron_variant | NA | NA | NA | NA | 1000Genomes,Cited,Frequency,gnomAD,Phenotype_or_Disease,TOPMed | ||||
| rs76904798 | LRRK2 | 12 | 40614434 | 0 | 0.1439 | 0.0130 | 0.1444 | 0.1444 | 56306 | 1417791 | 0.0382 | T | C | TRUE | NA | NA | 1 | 1 | 1 | 1 | 1 | 1 | 40.61443 | 3 | TRUE | 0.75 | 0 | 216621 | 11.069231 | 12 | 40220632 | 0 | 0 | 0 | C | T | 0.11 | 0.21 | 0.03 | 0.13 | 0 | 0 | E029,7_Enh;E030,7_Enh;E124,7_Enh | E029,H3K4me1_Enh;E030,H3K4me1_Enh;E031,H3K4me1_Enh;E124,H3K4me1_Enh;E124,H3K27ac_Enh | . | GTEx2015_v6,Adipose_Subcutaneous,LRRK2,2.6136557196757e-09;GTEx2015_v6,Artery_Coronary,LRRK2,8.10828434519411e-08;GTEx2015_v6,Artery_Tibial,LRRK2,1.29589062525083e-14;GTEx2015_v6,Cells_Transformed_fibroblasts,LRRK2,2.01546552400868e-05;GTEx2015_v6,Esophagus_Muscularis,LRRK2,1.17034509288903e-05;GTEx2015_v6,Nerve_Tibial,LRRK2,1.17833931025132e-15;GTEx2015_v6,Skin_Not_Sun_Exposed_Suprapubic,LRRK2,3.02678179339592e-06;GTEx2015_v6,Skin_Sun_Exposed_Lower_leg,LRRK2,1.21658440247309e-07 | 25064009,Parkinson’s disease,5E-14 | . | Dbx1;Hmx_2;Hoxb8;Hoxd8;Ncx_2;Nkx6-1_3;Pou2f2_known2;Sox_14;Sox_15;Sox_16;Sox_18;Sox_19;Sox_2;Sox_5;Sox_7;Sox_9 | ENSG00000225342.1 | AC079630.4 | 0 | 0 | NM_198578 | LRRK2 | 5 | 4377 | . | rs76904798 | BLD | 40614433 | 40614434 | FALSE | FALSE | FALSE | FALSE | 0 | 0.230000004172325 | TRUE | FALSE | FALSE | 0.14464 | 6 | C/T | 12 | 40614434 | 40614434 | 1 | ENSG00000188906 | intron_variant | NA | NA | NA | NA | 1000Genomes,Cited,Frequency,gnomAD,Phenotype_or_Disease,TOPMed |
Summary plots
Credible Set bin plot
gg_cs_bin <- echoannot::CS_bin_plot(merged_DT = merged_DT,
show_plot = FALSE)Credible Set counts plot
gg_cs_counts <- echoannot::CS_counts_plot(merged_DT = merged_DT,
show_plot = FALSE)Epigenomic data
gg_epi <- echoannot::peak_overlap_plot(
merged_DT = merged_DT,
include.NOTT2019_enhancers_promoters = TRUE,
include.NOTT2019_PLACseq = TRUE,
#### Omit many annotations to save time ####
include.NOTT2019_peaks = FALSE,
include.CORCES2020_scATACpeaks = FALSE,
include.CORCES2020_Cicero_coaccess = FALSE,
include.CORCES2020_bulkATACpeaks = FALSE,
include.CORCES2020_HiChIP_FitHiChIP_coaccess = FALSE,
include.CORCES2020_gene_annotations = FALSE)## ++ NOTT2019:: Getting regulatory regions data.## Importing Astrocyte enhancers ...## Importing Astrocyte promoters ...## Importing Neuronal enhancers ...## Importing Neuronal promoters ...## Importing Oligo enhancers ...## Importing Oligo promoters ...## Importing Microglia enhancers ...## Importing Microglia promoters ...## Converting dat to GRanges object.
## Converting dat to GRanges object.## 48 query SNP(s) detected with reference overlap.## ++ NOTT2019:: Getting interaction anchors data.## Importing Microglia interactome ...## Importing Neuronal interactome ...## Importing Oligo interactome ...## Converting dat to GRanges object.## 52 query SNP(s) detected with reference overlap.## Converting dat to GRanges object.## 44 query SNP(s) detected with reference overlap.## Found more than one class "simpleUnit" in cache; using the first, from namespace 'hexbin'## Also defined by 'ggbio'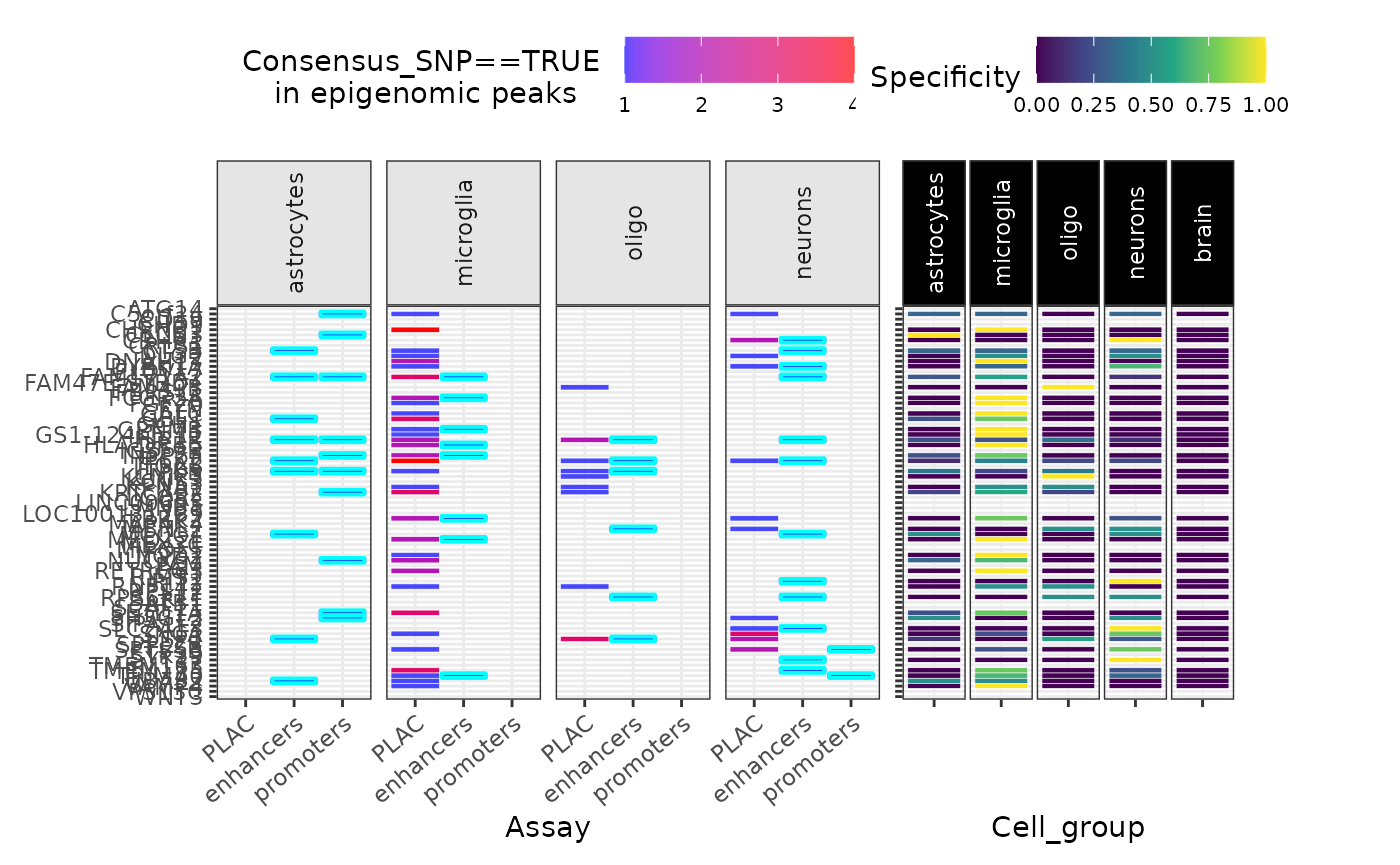
Super summary plot
Creates one big merged plot using the subfunctions above.
super_plot <- echoannot::super_summary_plot(merged_DT = merged_DT,
plot_missense = FALSE)Session Info
utils::sessionInfo()## R version 4.2.1 (2022-06-23)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 20.04.5 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/liblapack.so.3
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] echoannot_0.99.10 BiocStyle_2.25.0
##
## loaded via a namespace (and not attached):
## [1] utf8_1.2.2 reticulate_1.26
## [3] R.utils_2.12.0 RUnit_0.4.32
## [5] tidyselect_1.2.0 RSQLite_2.2.18
## [7] AnnotationDbi_1.59.1 htmlwidgets_1.5.4
## [9] grid_4.2.1 BiocParallel_1.31.15
## [11] XGR_1.1.8 munsell_0.5.0
## [13] codetools_0.2-18 ragg_1.2.4
## [15] interp_1.1-3 DT_0.26
## [17] withr_2.5.0 colorspace_2.0-3
## [19] OrganismDbi_1.39.1 Biobase_2.57.3
## [21] filelock_1.0.2 highr_0.9
## [23] knitr_1.40 supraHex_1.35.0
## [25] rstudioapi_0.14 stats4_4.2.1
## [27] DescTools_0.99.47 labeling_0.4.2
## [29] MatrixGenerics_1.9.1 GenomeInfoDbData_1.2.9
## [31] farver_2.1.1 bit64_4.0.5
## [33] echoconda_0.99.7 rprojroot_2.0.3
## [35] basilisk_1.9.12 vctrs_0.5.0
## [37] generics_0.1.3 xfun_0.34
## [39] biovizBase_1.45.0 BiocFileCache_2.5.2
## [41] R6_2.5.1 GenomeInfoDb_1.33.16
## [43] RJSONIO_1.3-1.6 AnnotationFilter_1.21.0
## [45] bitops_1.0-7 cachem_1.0.6
## [47] reshape_0.8.9 DelayedArray_0.23.2
## [49] assertthat_0.2.1 BiocIO_1.7.1
## [51] scales_1.2.1 nnet_7.3-18
## [53] rootSolve_1.8.2.3 gtable_0.3.1
## [55] lmom_2.9 ggbio_1.45.0
## [57] ensembldb_2.21.5 rlang_1.0.6
## [59] systemfonts_1.0.4 echodata_0.99.15
## [61] splines_4.2.1 lazyeval_0.2.2
## [63] rtracklayer_1.57.0 dichromat_2.0-0.1
## [65] hexbin_1.28.2 checkmate_2.1.0
## [67] BiocManager_1.30.19 yaml_2.3.6
## [69] reshape2_1.4.4 backports_1.4.1
## [71] GenomicFeatures_1.49.8 ggnetwork_0.5.10
## [73] Hmisc_4.7-1 RBGL_1.73.0
## [75] tools_4.2.1 bookdown_0.29
## [77] ggplot2_3.3.6 ellipsis_0.3.2
## [79] jquerylib_0.1.4 RColorBrewer_1.1-3
## [81] proxy_0.4-27 BiocGenerics_0.43.4
## [83] Rcpp_1.0.9 plyr_1.8.7
## [85] base64enc_0.1-3 progress_1.2.2
## [87] zlibbioc_1.43.0 purrr_0.3.5
## [89] RCurl_1.98-1.9 basilisk.utils_1.9.4
## [91] prettyunits_1.1.1 rpart_4.1.16
## [93] deldir_1.0-6 S4Vectors_0.35.4
## [95] cluster_2.1.4 SummarizedExperiment_1.27.3
## [97] ggrepel_0.9.1 fs_1.5.2
## [99] crul_1.3 magrittr_2.0.3
## [101] data.table_1.14.4 echotabix_0.99.8
## [103] dnet_1.1.7 openxlsx_4.2.5.1
## [105] mvtnorm_1.1-3 ProtGenerics_1.29.1
## [107] matrixStats_0.62.0 patchwork_1.1.2
## [109] hms_1.1.2 evaluate_0.17
## [111] XML_3.99-0.11 jpeg_0.1-9
## [113] readxl_1.4.1 IRanges_2.31.2
## [115] gridExtra_2.3 compiler_4.2.1
## [117] biomaRt_2.53.3 tibble_3.1.8
## [119] haploR_4.0.6 crayon_1.5.2
## [121] R.oo_1.25.0 htmltools_0.5.3
## [123] tzdb_0.3.0 Formula_1.2-4
## [125] tidyr_1.2.1 expm_0.999-6
## [127] Exact_3.2 DBI_1.1.3
## [129] dbplyr_2.2.1 MASS_7.3-58.1
## [131] rappdirs_0.3.3 boot_1.3-28
## [133] Matrix_1.5-1 readr_2.1.3
## [135] piggyback_0.1.4 cli_3.4.1
## [137] R.methodsS3_1.8.2 parallel_4.2.1
## [139] igraph_1.3.5 GenomicRanges_1.49.1
## [141] pkgconfig_2.0.3 pkgdown_2.0.6.9000
## [143] GenomicAlignments_1.33.1 dir.expiry_1.5.1
## [145] RCircos_1.2.2 foreign_0.8-83
## [147] osfr_0.2.9 xml2_1.3.3
## [149] bslib_0.4.0 XVector_0.37.1
## [151] stringr_1.4.1 VariantAnnotation_1.43.3
## [153] digest_0.6.30 graph_1.75.1
## [155] httpcode_0.3.0 Biostrings_2.65.6
## [157] rmarkdown_2.17 cellranger_1.1.0
## [159] htmlTable_2.4.1 gld_2.6.6
## [161] restfulr_0.0.15 curl_4.3.3
## [163] Rsamtools_2.13.4 rjson_0.2.21
## [165] lifecycle_1.0.3 nlme_3.1-160
## [167] jsonlite_1.8.3 viridisLite_0.4.1
## [169] desc_1.4.2 BSgenome_1.65.4
## [171] fansi_1.0.3 downloadR_0.99.5
## [173] pillar_1.8.1 lattice_0.20-45
## [175] GGally_2.1.2 KEGGREST_1.37.3
## [177] fastmap_1.1.0 httr_1.4.4
## [179] survival_3.4-0 glue_1.6.2
## [181] zip_2.2.2 png_0.1-7
## [183] bit_4.0.4 Rgraphviz_2.41.2
## [185] class_7.3-20 stringi_1.7.8
## [187] sass_0.4.2 blob_1.2.3
## [189] textshaping_0.3.6 latticeExtra_0.6-30
## [191] memoise_2.0.1 dplyr_1.0.10
## [193] e1071_1.7-12 ape_5.6-2