`Get Started
¶ Author:
Brian M. Schilder ¶
¶ Updated:
Dec-23-2022 ¶
Source: vignettes/echoplot.Rmd
echoplot.RmdPlotting loci with echoplot
echoplot contains various functions that can be used
separately
from the comprehensive echolocatoR::finemap_loci()
pipeline.
Generate a multi-view plot of a given locus using
echoplot::plot_locus().
- You can mix and match different tracks and annotations using the
different arguments (see
?plot_locusfor details).
The plot is centered on the lead/index SNP. If a list is supplied to
zoom * plot_locus() returns a series of ggplot
objects bound together with patchwork.
One can further modify this object using ggplot2 functions
like + theme(). + The modifications will be applied to all
tracks at once.
- Save a high-resolution versions the plot by setting
save_plot=T.- Further increase resolution by adjusting the
dpiargument (default=300). - Files are saved in jpg format by default, but users can
specify their preferred file format
(e.g.
file_format="png") - Adjust the
heightandwidthof the saved plot using these respective arguments. - The plot will be automatically saved in the locus-specific directory
as:
*multiview__ .jpg*.
- Further increase resolution by adjusting the
Load example data
Load example dataset of the results from fine-mapping the BST1 locus
with finemap_loci(). Original data comes from the recent
Nalls et al. (2019) Parkinson’s disease GWAS (see ?BST1 for
details).
Full window
trk_plot <- echoplot:: plot_locus(dat=dat,
LD_matrix=LD_matrix,
LD_reference=LD_reference,
locus_dir=locus_dir,
save_plot=FALSE,
show_plot=show_plot,
zoom=zoom) ## +-------- Locus Plot: BST1 --------+## + support_thresh = 2## + Calculating mean Posterior Probability (mean.PP)...## + 4 fine-mapping methods used.## + 7 Credible Set SNPs identified.## + 3 Consensus SNPs identified.## + Filling NAs in CS cols with 0.## + Filling NAs in PP cols with 0.## LD_matrix detected. Coloring SNPs by LD with lead SNP.## Filling r/r2 NAs with 0## ++ echoplot:: GWAS full window track## ++ echoplot:: GWAS track## ++ echoplot:: Merged fine-mapping track## Melting PP and CS from 5 fine-mapping methods.## + echoplot:: Constructing SNP labels.## Adding SNP group labels to locus plot.## ++ echoplot:: Adding Gene model track.## Converting dat to GRanges object.## Loading required namespace: EnsDb.Hsapiens.v75## max_transcripts= 1 .## 16 transcripts from 16 genes returned.## Fetching data...OK
## Parsing exons...OK
## Defining introns...OK
## Defining UTRs...OK
## Defining CDS...OK
## aggregating...
## Done
## Constructing graphics...
## + Adding vertical lines to highlight SNP groups.
## +>+>+>+>+ zoom = 10x +<+<+<+<+
## + echoplot:: Get window suffix...
## + Constructing zoom polygon...
## + Highlighting zoom origin...
## + Removing subplot margins...
## + Reordering tracks...
## + Ensuring last track shows genomic units.
## + Aligning xlimits for each subplot...
## + Checking track heights...
methods::show(trk_plot)## $`10x`## Found more than one class "simpleUnit" in cache; using the first, from namespace 'hexbin'## Also defined by 'ggbio'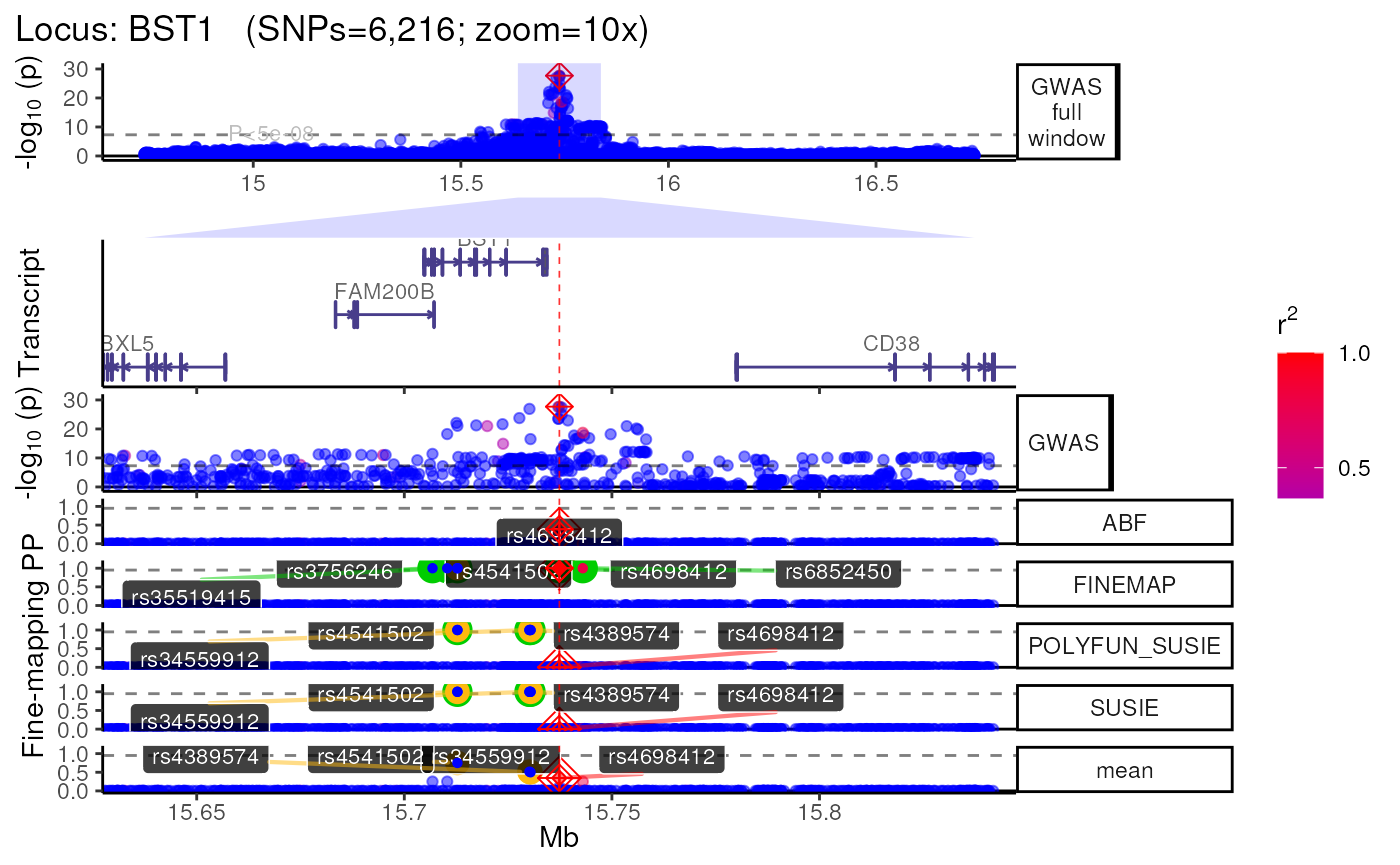
At multiple zooms
- You can easily generate the same locus plot at multiple zoomed in
views by supplying a list to
zoom.
- This list can be composed of zoom multipliers
(e.g.
c("1x", "2x")), window widths in units of basepairs (e.g.c(5000, 1500)), or a mixture of both (e.g.c("1x","4x", 5000, 2000)). - Each zoom view will be saved individually with its respective scale
as the suffix (e.g.
multiview.BST1.UKB.4x.jpg).
- Each zoom view is stored as a named item within the returned list.
trk_zooms <- plot_locus(dat=dat,
LD_matrix=LD_matrix,
LD_reference=LD_reference,
locus_dir=locus_dir,
save_plot=FALSE,
show_plot=show_plot,
zoom = c("1x","5x","10x")) ## +-------- Locus Plot: BST1 --------+## + support_thresh = 2## + Calculating mean Posterior Probability (mean.PP)...## + 4 fine-mapping methods used.## + 7 Credible Set SNPs identified.## + 3 Consensus SNPs identified.## + Filling NAs in CS cols with 0.## + Filling NAs in PP cols with 0.## LD_matrix detected. Coloring SNPs by LD with lead SNP.## Filling r/r2 NAs with 0## ++ echoplot:: GWAS full window track## ++ echoplot:: GWAS track## ++ echoplot:: Merged fine-mapping track## Melting PP and CS from 5 fine-mapping methods.## + echoplot:: Constructing SNP labels.## Adding SNP group labels to locus plot.## ++ echoplot:: Adding Gene model track.## Converting dat to GRanges object.## max_transcripts= 1 .## 16 transcripts from 16 genes returned.## Fetching data...OK
## Parsing exons...OK
## Defining introns...OK
## Defining UTRs...OK
## Defining CDS...OK
## aggregating...
## Done
## Constructing graphics...
## + Adding vertical lines to highlight SNP groups.
## +>+>+>+>+ zoom = 1x +<+<+<+<+
## + echoplot:: Get window suffix...
## + echoplot:: Removing GWAS full window track @ zoom=1x
## + Removing subplot margins...
## + Reordering tracks...
## + Ensuring last track shows genomic units.
## + Aligning xlimits for each subplot...
## + Checking track heights...
## +>+>+>+>+ zoom = 5x +<+<+<+<+
## + echoplot:: Get window suffix...
## + Constructing zoom polygon...
## + Highlighting zoom origin...
## + Removing subplot margins...
## + Reordering tracks...
## + Ensuring last track shows genomic units.
## + Aligning xlimits for each subplot...
## + Checking track heights...
## +>+>+>+>+ zoom = 10x +<+<+<+<+
## + echoplot:: Get window suffix...
## + Constructing zoom polygon...
## + Highlighting zoom origin...
## + Removing subplot margins...
## + Reordering tracks...
## + Ensuring last track shows genomic units.
## + Aligning xlimits for each subplot...
## + Checking track heights...
names(trk_zooms) # Get zoom view names## [1] "1x" "5x" "10x"
methods::show(trk_zooms)## $`1x`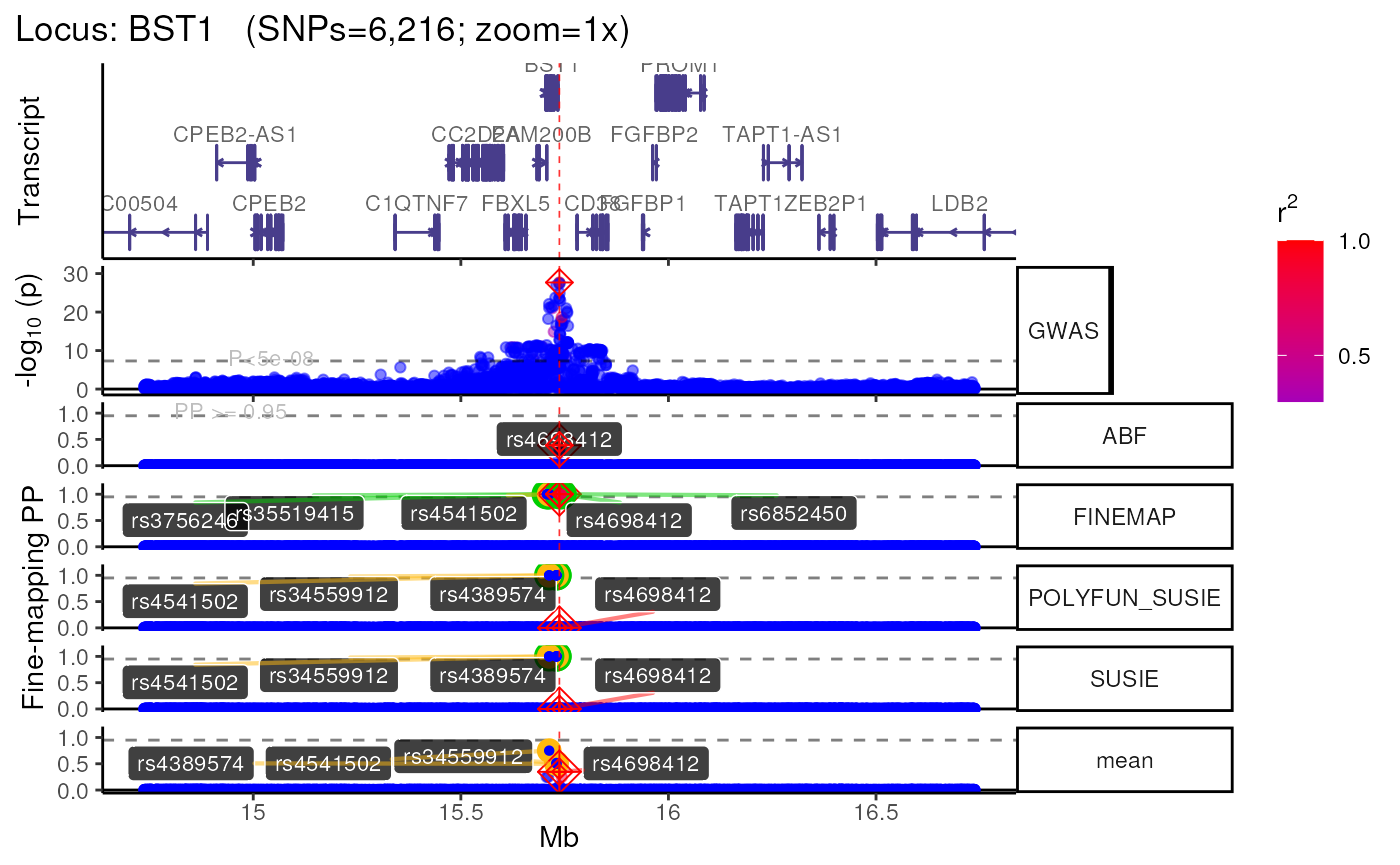
##
## $`5x`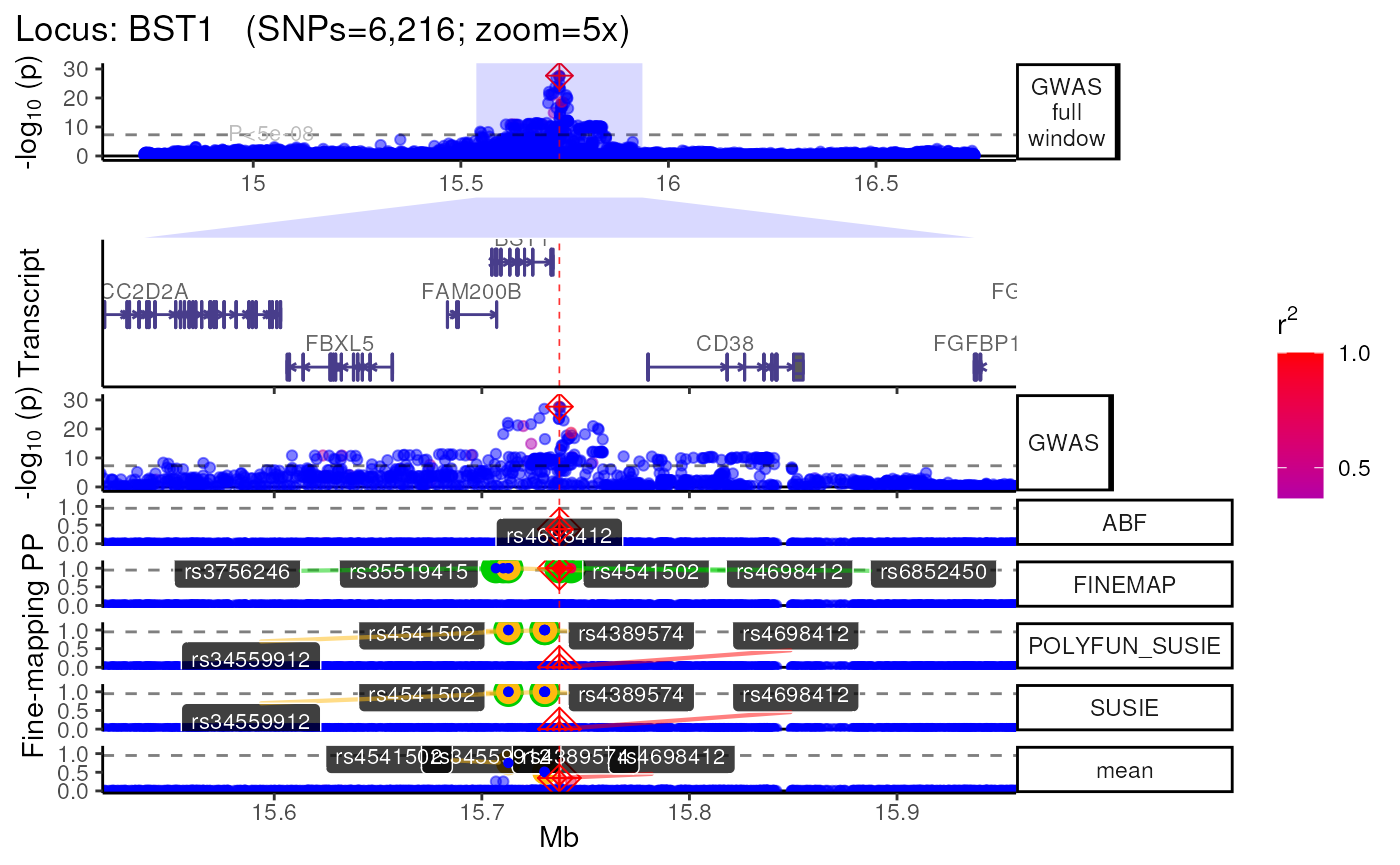
##
## $`10x`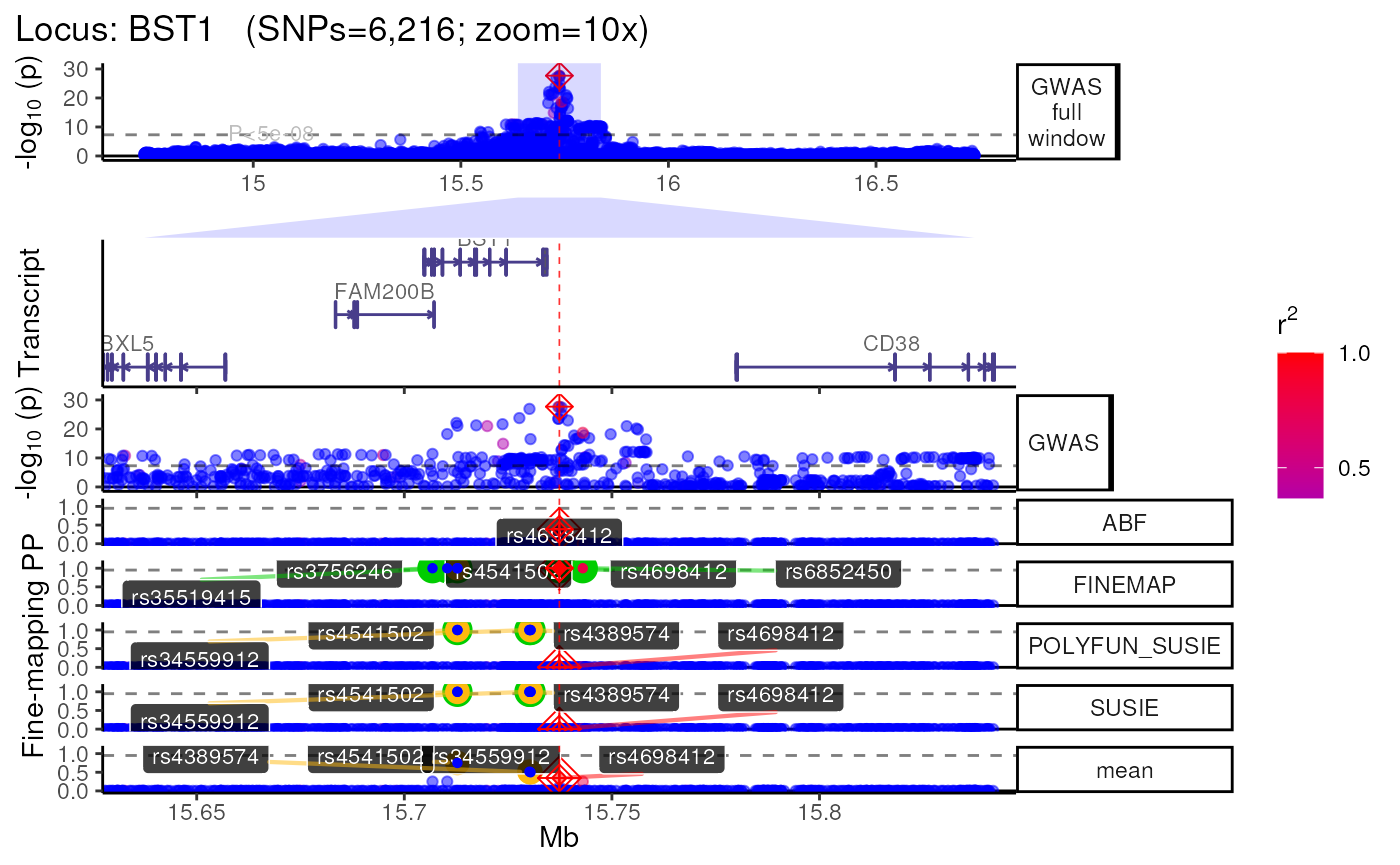
Return as list
- For even further control over each track of the multi-view plot,
specify
plot_locus(..., return_list=TRUE)to instead return a named list (nested within each zoom view list item) ofggplotobjects which can each be modified individually. - Once you’ve made your modifications, you can then bind this list of
plots back together with
patchwork::wrap_plots(tracks_list, ncol = 1).
trk_plot_list <- echoplot::plot_locus(dat=dat,
LD_matrix=LD_matrix,
LD_reference=LD_reference,
locus_dir=locus_dir,
save_plot=FALSE,
show_plot=show_plot,
zoom=zoom,
return_list=TRUE) ## +-------- Locus Plot: BST1 --------+## + support_thresh = 2## + Calculating mean Posterior Probability (mean.PP)...## + 4 fine-mapping methods used.## + 7 Credible Set SNPs identified.## + 3 Consensus SNPs identified.## + Filling NAs in CS cols with 0.## + Filling NAs in PP cols with 0.## LD_matrix detected. Coloring SNPs by LD with lead SNP.## Filling r/r2 NAs with 0## ++ echoplot:: GWAS full window track## ++ echoplot:: GWAS track## ++ echoplot:: Merged fine-mapping track## Melting PP and CS from 5 fine-mapping methods.## + echoplot:: Constructing SNP labels.## Adding SNP group labels to locus plot.## ++ echoplot:: Adding Gene model track.## Converting dat to GRanges object.## max_transcripts= 1 .## 16 transcripts from 16 genes returned.## Fetching data...OK
## Parsing exons...OK
## Defining introns...OK
## Defining UTRs...OK
## Defining CDS...OK
## aggregating...
## Done
## Constructing graphics...
## + Adding vertical lines to highlight SNP groups.
## +>+>+>+>+ zoom = 10x +<+<+<+<+
## + echoplot:: Get window suffix...
## + Constructing zoom polygon...
## + Highlighting zoom origin...
## + Removing subplot margins...
## + Reordering tracks...
## + Ensuring last track shows genomic units.
## + Aligning xlimits for each subplot...
## + Checking track heights...
view1_list <- trk_plot_list[[zoom]]
names(view1_list) # Get track names from a particular zoom view## [1] "GWAS full window" "zoom_polygon" "Genes" "GWAS"
## [5] "Fine-mapping"Modify a specific tracks within a view.
# Modify your selected track
modified_track <- view1_list$GWAS +
ggplot2::labs(title = "Modified GWAS") +
ggplot2::theme_dark() +
ggplot2::theme(title = ggplot2::element_text(hjust = .5))
# Put it back into your track list
view1_list[["GWAS"]] <- modified_track
# Remove a plot you don't want
view1_list[["Genes"]] <- NULL
# Specify the relative heights of each track (make sure it matches your new # of plots!)
track_heights <- c(.3,.1,.3,1)
# Bind them together and plot
fused_plot <- patchwork::wrap_plots(view1_list,
heights = track_heights,
ncol = 1)
methods::show(fused_plot)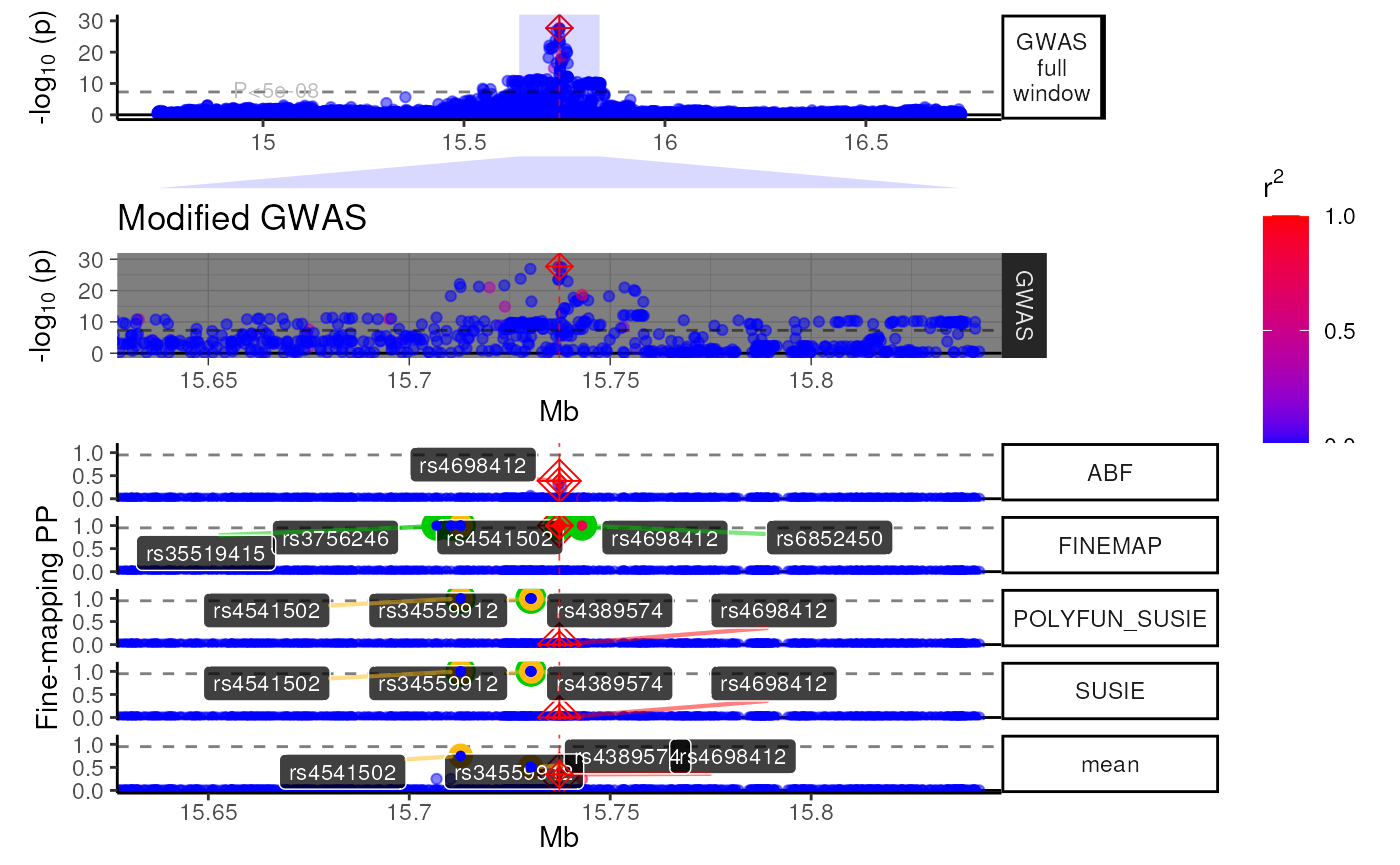
Using XGR annotations
- Whenever you use annotation arguments
(e.g.
xgr_libnames,Roadmap,nott_epigenome) the annotations that overlap with your locus will automatically be saved asGRangesobjects in a locus-specific subdirectory:
results// / /annotation - If a selected annotation has previously been downloaded and stored
for that locus,
plot_locus()will automatically detect and import it to save time.
trk_plot.xgr <- echoplot::plot_locus(dat=dat,
LD_matrix=LD_matrix,
LD_reference=LD_reference,
locus_dir=locus_dir,
xgr_libnames=c("ENCODE_TFBS_ClusteredV3_CellTypes"),
save_plot=FALSE,
show_plot=show_plot,
zoom=zoom)## +-------- Locus Plot: BST1 --------+## + support_thresh = 2## + Calculating mean Posterior Probability (mean.PP)...## + 4 fine-mapping methods used.## + 7 Credible Set SNPs identified.## + 3 Consensus SNPs identified.## + Filling NAs in CS cols with 0.## + Filling NAs in PP cols with 0.## LD_matrix detected. Coloring SNPs by LD with lead SNP.## Filling r/r2 NAs with 0## ++ echoplot:: GWAS full window track## ++ echoplot:: GWAS track## ++ echoplot:: Merged fine-mapping track## Melting PP and CS from 5 fine-mapping methods.## + echoplot:: Constructing SNP labels.## Adding SNP group labels to locus plot.## ++ echoplot:: Adding Gene model track.## Converting dat to GRanges object.## max_transcripts= 1 .## 16 transcripts from 16 genes returned.## Fetching data...OK
## Parsing exons...OK
## Defining introns...OK
## Defining UTRs...OK
## Defining CDS...OK
## aggregating...
## Done
## Constructing graphics...
## echoannot:: Plotting XGR annotations.
## Start at 2022-12-23 02:23:34.092123
##
## 'ENCODE_TFBS_ClusteredV3_CellTypes' (from http://galahad.well.ox.ac.uk/bigdata/ENCODE_TFBS_ClusteredV3_CellTypes.RData) has been loaded into the working environment (at 2022-12-23 02:23:42.436726)
##
## End at 2022-12-23 02:23:42.437574
## Runtime in total is: 8 secs
##
## Converting dat to GRanges object.
## 1,579 query SNP(s) detected with reference overlap.## Warning in (function (mapping = NULL, data = NULL, stat = "density", position =
## "identity", : Ignoring unknown parameters: `facets`## Warning in max(xlim): no non-missing arguments to max; returning -Inf## + Adding vertical lines to highlight SNP groups.## Warning: Groups with fewer than two data points have been dropped.## Warning: Groups with fewer than two data points have been dropped.## Warning: Removed 2 rows containing missing values (`position_stack()`).## +>+>+>+>+ zoom = 10x +<+<+<+<+
## + echoplot:: Get window suffix...
## + Constructing zoom polygon...
## + Highlighting zoom origin...
## + Removing subplot margins...
## + Reordering tracks...
## + Ensuring last track shows genomic units.## Warning: Groups with fewer than two data points have been dropped.## Warning: Groups with fewer than two data points have been dropped.## Warning: Removed 2 rows containing missing values (`position_stack()`).## + Aligning xlimits for each subplot...## Warning: Groups with fewer than two data points have been dropped.## Warning: Groups with fewer than two data points have been dropped.## Warning: Removed 2 rows containing missing values (`position_stack()`).## + Checking track heights...
methods::show(trk_plot.xgr)## $`10x`## Warning: Groups with fewer than two data points have been dropped.
## Groups with fewer than two data points have been dropped.## Warning: Removed 2 rows containing missing values (`position_stack()`).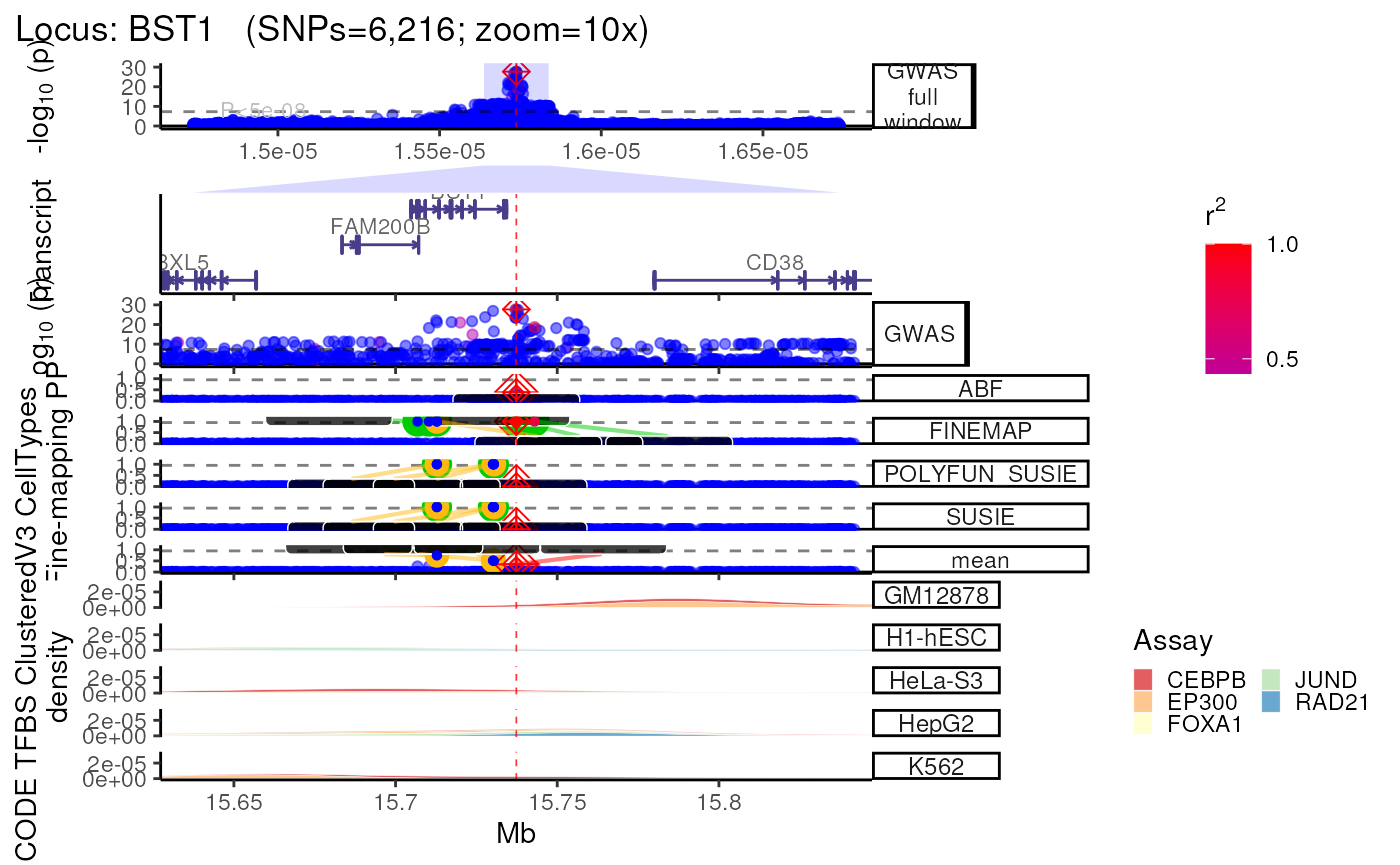
Using Roadmap annotations
- Using the
Roadmap=Tandroadmap_query="<query>"arguments searches the Roadmap for chromatin mark data across various cell-types, cell-lines and tissues.
- Note that Roadmap queries requires
tabixto be installed on your machine, or within a conda environment (conda_env = "echoR"). - Parallelizing these queries across multiple thredas speeds up this
process (
nThread=<n_cores_available>), as does reusing previously stored data which is automatically saved to the locus-specific subfolder (<dataset_type>/<dataset_name>/<locus>/annotations/Roadmap.ChromatinMarks_CellTypes.RDS).
trk_plot.roadmap <- echoplot::plot_locus(dat=dat,
LD_matrix=LD_matrix,
LD_reference=LD_reference,
locus_dir=locus_dir,
roadmap=TRUE,
roadmap_query="monocyte",
save_plot=FALSE,
show_plot=show_plot,
zoom="5x")## +-------- Locus Plot: BST1 --------+## + support_thresh = 2## + Calculating mean Posterior Probability (mean.PP)...## + 4 fine-mapping methods used.## + 7 Credible Set SNPs identified.## + 3 Consensus SNPs identified.## + Filling NAs in CS cols with 0.## + Filling NAs in PP cols with 0.## LD_matrix detected. Coloring SNPs by LD with lead SNP.## Filling r/r2 NAs with 0## ++ echoplot:: GWAS full window track## ++ echoplot:: GWAS track## ++ echoplot:: Merged fine-mapping track## Melting PP and CS from 5 fine-mapping methods.## + echoplot:: Constructing SNP labels.## Adding SNP group labels to locus plot.## ++ echoplot:: Adding Gene model track.## Converting dat to GRanges object.## max_transcripts= 1 .## 16 transcripts from 16 genes returned.## Fetching data...OK
## Parsing exons...OK
## Defining introns...OK
## Defining UTRs...OK
## Defining CDS...OK
## aggregating...
## Done
## Constructing graphics...
## echoannot:: Plotting ROADMAP annotations.
## Converting dat to GRanges object.
## + ROADMAP:: 2 annotation(s) identified that match: monocyte
## Constructing GRanges query using min/max ranges across one or more chromosomes.
## + as_blocks=TRUE: Will query a single range per chromosome that covers all regions requested (plus anything in between).
## Downloading Roadmap Chromatin Marks: E124
## Converting dat to GRanges object.
## Saving query ==> /tmp/RtmpD97Fpx/roadmap_query--monocyte.n_top--7.limit_files--./ROADMAP_query.E124.rds
## Converting 1 GRanges object to separate BED files.
## Saving BED file ==> /tmp/RtmpD97Fpx/roadmap_query--monocyte.n_top--7.limit_files--./E124.bed.txt.gz
## BED subset downloaded in 8.555 seconds
## Constructing GRanges query using min/max ranges across one or more chromosomes.
## + as_blocks=TRUE: Will query a single range per chromosome that covers all regions requested (plus anything in between).
## Downloading Roadmap Chromatin Marks: E029
## Converting dat to GRanges object.
## Saving query ==> /tmp/RtmpD97Fpx/roadmap_query--monocyte.n_top--7.limit_files--./ROADMAP_query.E029.rds
## Converting 1 GRanges object to separate BED files.
## Saving BED file ==> /tmp/RtmpD97Fpx/roadmap_query--monocyte.n_top--7.limit_files--./E029.bed.txt.gz
## BED subset downloaded in 8.517 seconds
## ROADMAP:: Annotating GRangesList.
## Annotating chromatin states.
## Merging and processing ROADMAP annotations.
## ROADMAP:: Done in 0.32 min.
## Generating ROADMAP track plot.## Warning in (function (mapping = NULL, data = NULL, stat = "density", position =
## "identity", : Ignoring unknown parameters: `facets`## Warning in max(xlim): no non-missing arguments to max; returning -Inf## + Adding vertical lines to highlight SNP groups.## Warning: Groups with fewer than two data points have been dropped.## Warning: Groups with fewer than two data points have been dropped.
## Groups with fewer than two data points have been dropped.
## Groups with fewer than two data points have been dropped.## Warning: Removed 4 rows containing missing values (`position_stack()`).## +>+>+>+>+ zoom = 5x +<+<+<+<+
## + echoplot:: Get window suffix...
## + Constructing zoom polygon...
## + Highlighting zoom origin...
## + Removing subplot margins...
## + Reordering tracks...
## + Ensuring last track shows genomic units.## Warning: Groups with fewer than two data points have been dropped.## Warning: Groups with fewer than two data points have been dropped.
## Groups with fewer than two data points have been dropped.
## Groups with fewer than two data points have been dropped.## Warning: Removed 4 rows containing missing values (`position_stack()`).## + Aligning xlimits for each subplot...## Warning: Groups with fewer than two data points have been dropped.## Warning: Groups with fewer than two data points have been dropped.
## Groups with fewer than two data points have been dropped.
## Groups with fewer than two data points have been dropped.## Warning: Removed 4 rows containing missing values (`position_stack()`).## + Checking track heights...
methods::show(trk_plot.roadmap)## $`5x`## Warning: Groups with fewer than two data points have been dropped.
## Groups with fewer than two data points have been dropped.
## Groups with fewer than two data points have been dropped.
## Groups with fewer than two data points have been dropped.## Warning: Removed 4 rows containing missing values (`position_stack()`).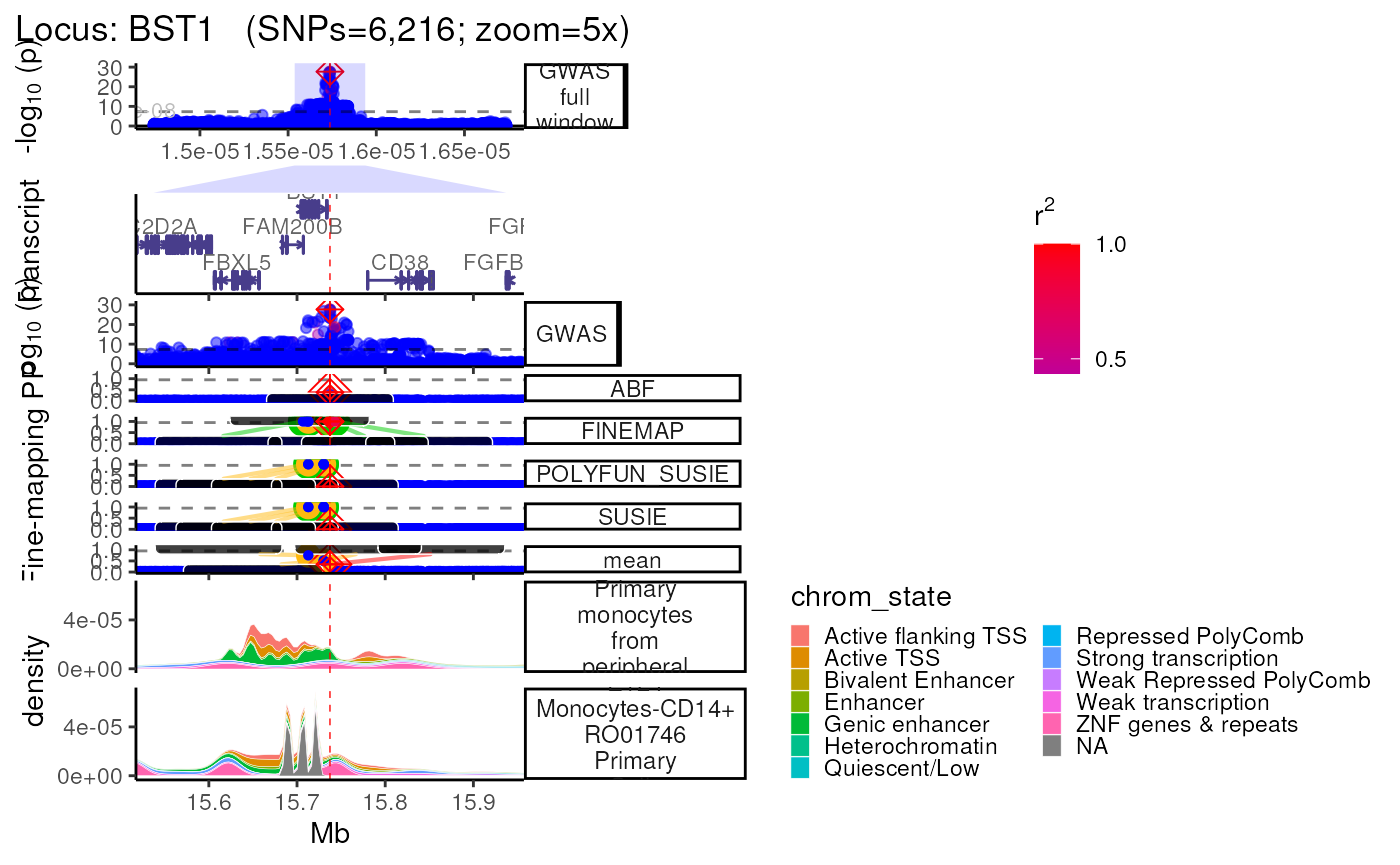
Using Nott_2019 annotations
- Query and plot brain cell type-specific epigenomic assays from Nott
et al. (Science, 2019)
(see?NOTT_2019.bigwig_metadatafor details).
trk_plot.nott_2019 <- echoplot::plot_locus(dat=dat,
LD_matrix=LD_matrix,
LD_reference=LD_reference,
locus_dir=locus_dir,
nott_epigenome=TRUE,
nott_binwidth = 200,
nott_regulatory_rects = TRUE,
nott_show_placseq = TRUE,
save_plot=FALSE,
show_plot=show_plot,
zoom=zoom) ## +-------- Locus Plot: BST1 --------+## + support_thresh = 2## + Calculating mean Posterior Probability (mean.PP)...## + 4 fine-mapping methods used.## + 7 Credible Set SNPs identified.## + 3 Consensus SNPs identified.## + Filling NAs in CS cols with 0.## + Filling NAs in PP cols with 0.## LD_matrix detected. Coloring SNPs by LD with lead SNP.## Filling r/r2 NAs with 0## ++ echoplot:: GWAS full window track## ++ echoplot:: GWAS track## ++ echoplot:: Merged fine-mapping track## Melting PP and CS from 5 fine-mapping methods.## + echoplot:: Constructing SNP labels.## Adding SNP group labels to locus plot.## ++ echoplot:: Adding Gene model track.## Converting dat to GRanges object.## max_transcripts= 1 .## 16 transcripts from 16 genes returned.## Fetching data...OK
## Parsing exons...OK
## Defining introns...OK
## Defining UTRs...OK
## Defining CDS...OK
## aggregating...
## Done
## Constructing graphics...
## NOTT2019:: Creating epigenomic histograms plot
## + Inferring genomic limits for window: 1x
## Constructing GRanges query using min/max ranges across one or more chromosomes.
## Downloading data from UCSC.
## Importing... [1] exvivo_H3K27ac_tbp
## Importing... [2] microglia_H3K27ac
## Importing... [3] neurons_H3K27ac
## Importing... [4] oligodendrocytes_H3K27ac
## Importing... [5] astrocytes_H3K27ac
## Importing... [6] exvivo_atac_tbp
## Importing... [7] microglia_atac
## Importing... [8] neurons_atac
## Importing... [9] oligodendrocytes_atac
## Importing... [10] astrocytes_atac
## Importing... [11] microglia_H3K4me3
## Importing... [12] neurons_H3K4me3
## Importing... [13] oligodendrocytes_H3K4me3
## Importing... [14] astrocytes_H3K4me3
## Saving bigwig query ==> /tmp/RtmpD97Fpx/BST1_Nott2019_bigwig.rds
## Importing previously downloaded files: /github/home/.cache/R/echoannot/NOTT2019_epigenomic_peaks.rds
## ++ NOTT2019:: 634,540 ranges retrieved.
## dat is already a GRanges object.
## 543 query SNP(s) detected with reference overlap.
## + Calculating max histogram height
## + Converting label units to Mb.
## NOTT2019:: Creating PLAC-seq interactome plot
## ++ NOTT2019:: Getting promoter cell-type-specific data.
## ++ NOTT2019:: Getting interactome data.
## ++ NOTT2019:: Getting regulatory regions data.
## Importing Astrocyte enhancers ...
## Importing Astrocyte promoters ...
## Importing Neuronal enhancers ...
## Importing Neuronal promoters ...
## Importing Oligo enhancers ...
## Importing Oligo promoters ...
## Importing Microglia enhancers ...
## Importing Microglia promoters ...
## Converting dat to GRanges object.
## ++ NOTT2019:: Getting interaction anchors data.
## Importing Microglia interactome ...
## Importing Neuronal interactome ...
## Importing Oligo interactome ...
## Converting dat to GRanges object.
## 29 query SNP(s) detected with reference overlap.
## Converting dat to GRanges object.
## 49 query SNP(s) detected with reference overlap.
## Converting dat to GRanges object.
## Preparing data for highlighting PLAC-seq interactions that overlap with SNP subset: Support>0
## Saving annotations ==> /tmp/RtmpD97Fpx/results/GWAS/Nalls23andMe_2019/BST1/annotations/NOTT2019_interactome.rds
## Saving annotation ==> /tmp/RtmpD97Fpx/results/GWAS/Nalls23andMe_2019/BST1/annotations/NOTT2019_enhancers_promoters.rds
## Initializing PLAC-seq plot.
## ++ Adding enhancer/promoter rectangles
## ++ Removing xtext.
## x_limits will be used to limit the min/max x-axis values for all plots.
## Converting plots to a named list of ggplot objects.
## + Adding vertical lines to highlight SNP groups.
## +>+>+>+>+ zoom = 10x +<+<+<+<+
## + echoplot:: Get window suffix...
## + Constructing zoom polygon...
## + Highlighting zoom origin...
## + Removing subplot margins...
## + Reordering tracks...
## + Ensuring last track shows genomic units.
## + Aligning xlimits for each subplot...
## + Checking track heights...
methods::show(trk_plot.nott_2019)## $`10x`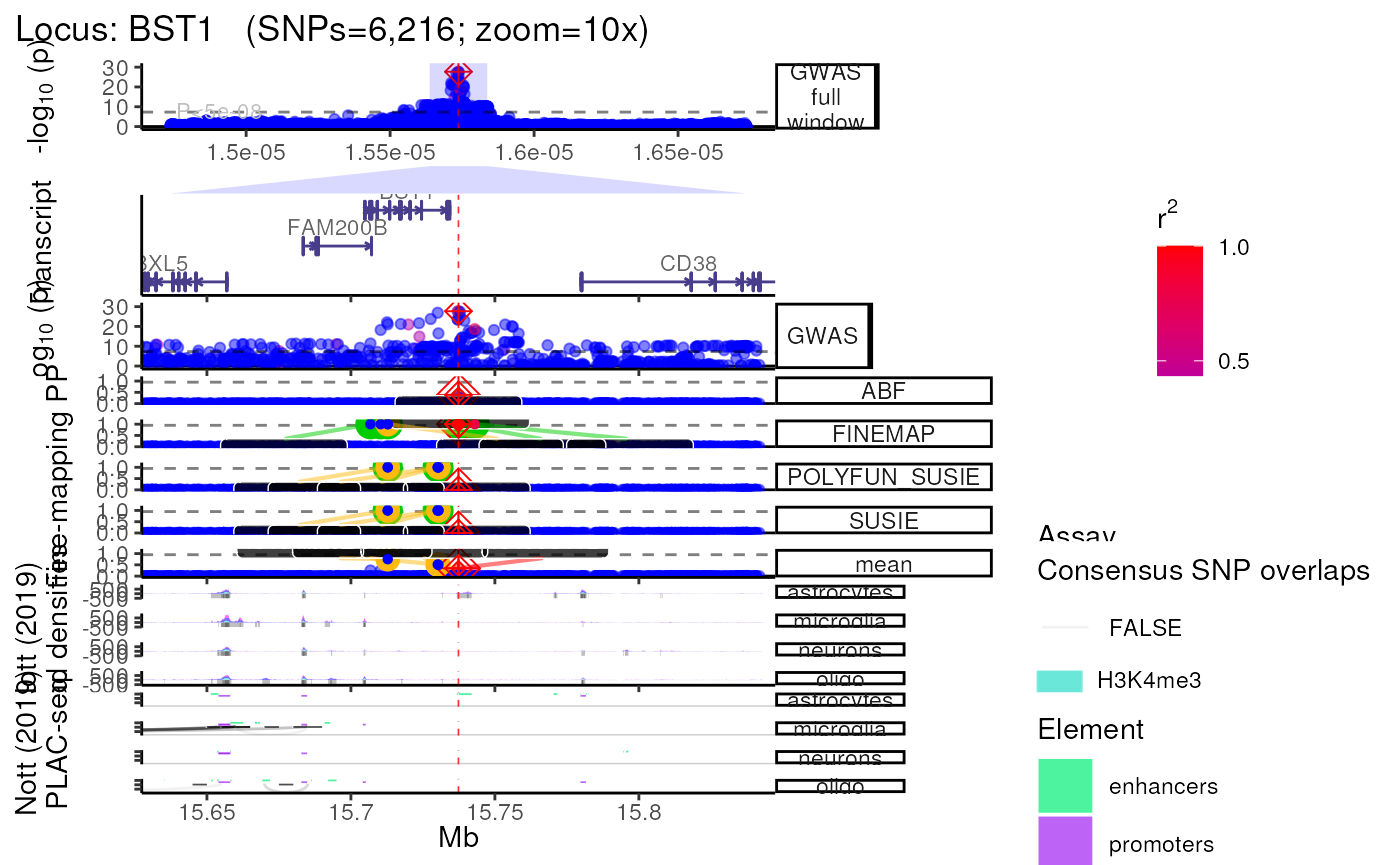
Using QTL datasets
- Plot multiple QTL p-value columns (or really P-value columns from
any kind of dataset).
- Each QTL dataset will be plotted as a new track.
dat1 <- data.table::copy(dat)
dat2 <- data.table::copy(dat)
# Make fake QTL P-values for the sake a demonstration
dat1$P <- abs(jitter(dat1$P, amount = 1e-15))
dat2$P <- abs(jitter(dat2$P, amount = 1e-16))
dat_ls <- list("fake_eQTL"=dat1,
"fake_sQTL"=dat2)
trk_plot.qtl <- echoplot::plot_locus_multi(dat_ls = dat_ls,
LD_ls = list(LD_matrix,LD_matrix),
locus_dir = locus_dir,
show_plot = show_plot,
zoom = "10x")## LD_matrix detected. Coloring SNPs by LD with lead SNP.## Filling r/r2 NAs with 0## LD_matrix detected. Coloring SNPs by LD with lead SNP.## Filling r/r2 NAs with 0## +-------- Locus Plot: BST1 --------+## + support_thresh = 2## + Calculating mean Posterior Probability (mean.PP)...## + 4 fine-mapping methods used.## + 14 Credible Set SNPs identified.## + 6 Consensus SNPs identified.## + Filling NAs in CS cols with 0.## + Filling NAs in PP cols with 0.## ++ echoplot:: GWAS full window track## ++ echoplot:: GWAS track## ++ echoplot:: Merged fine-mapping track## Melting PP and CS from 5 fine-mapping methods.## + echoplot:: Constructing SNP labels.## Adding SNP group labels to locus plot.## ++ echoplot:: Adding Gene model track.## Converting dat to GRanges object.## max_transcripts= 1 .## 16 transcripts from 16 genes returned.## Fetching data...OK
## Parsing exons...OK
## Defining introns...OK
## Defining UTRs...OK
## Defining CDS...OK
## aggregating...
## Done
## Constructing graphics...
## + Adding vertical lines to highlight SNP groups.
## +>+>+>+>+ zoom = 10x +<+<+<+<+
## + echoplot:: Get window suffix...
## + Constructing zoom polygon...
## + Highlighting zoom origin...
## + Removing subplot margins...
## + Reordering tracks...
## + Ensuring last track shows genomic units.
## + Aligning xlimits for each subplot...
## + Checking track heights...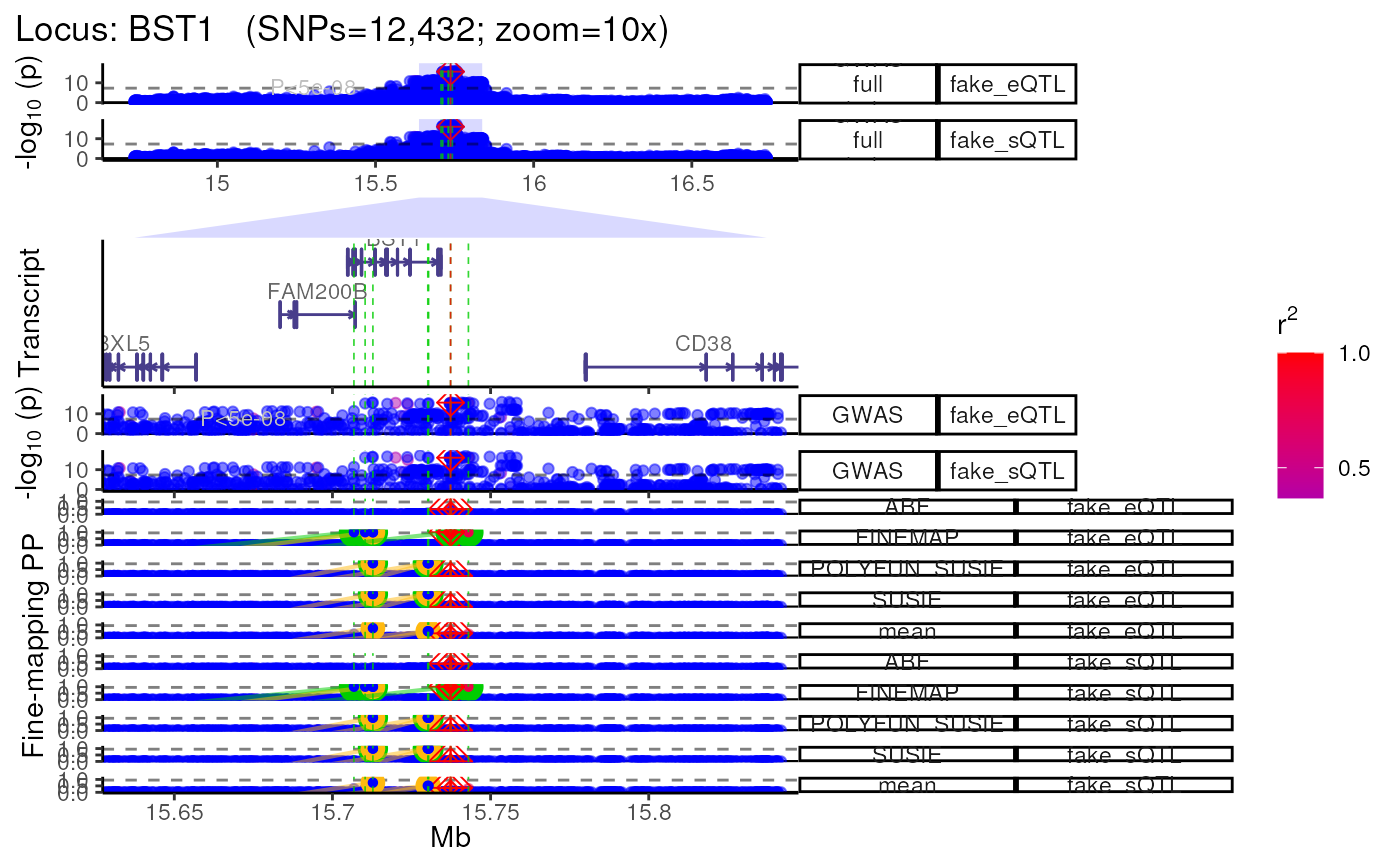
methods::show(trk_plot.qtl)## $`10x`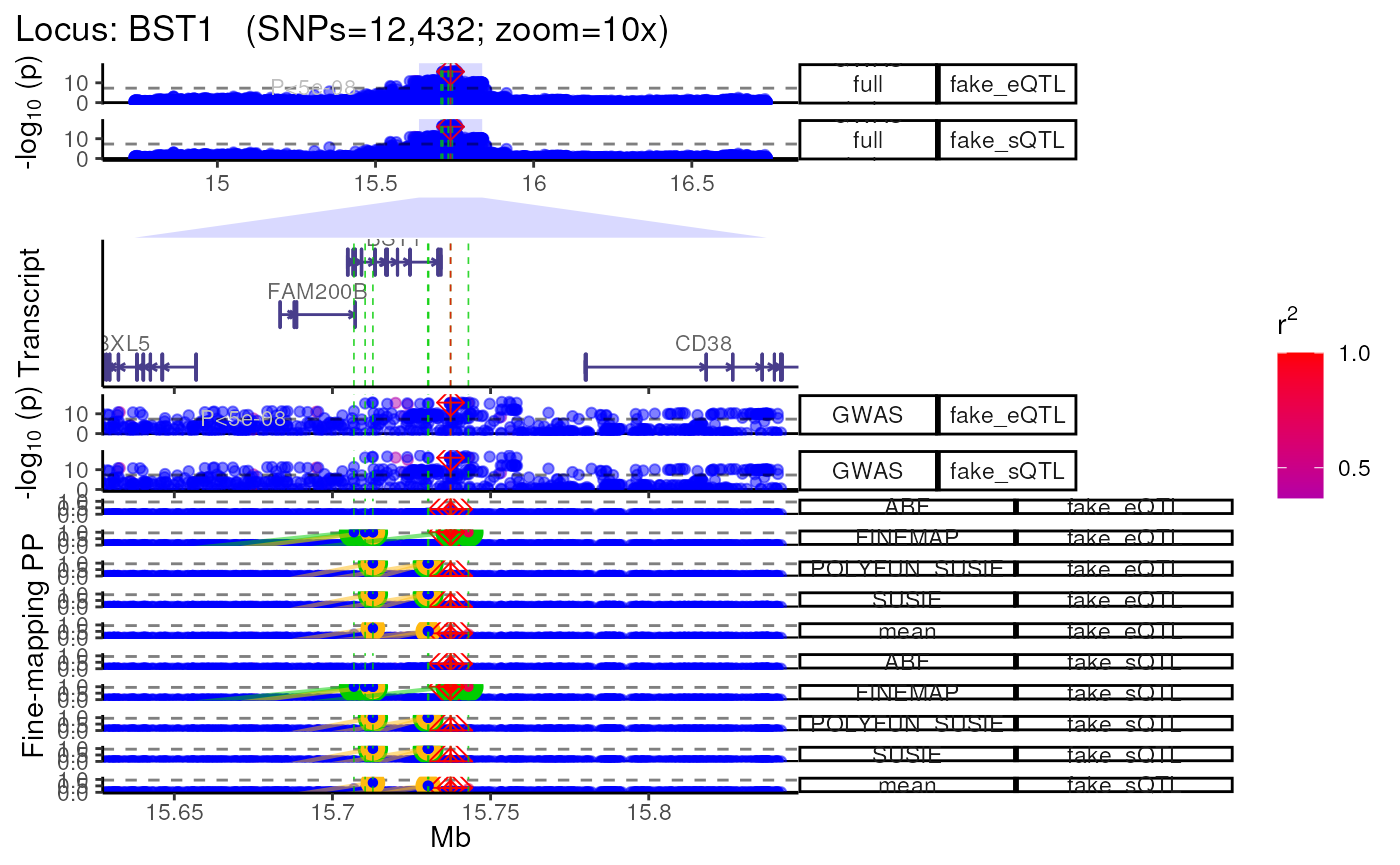
Session info
utils::sessionInfo()## R Under development (unstable) (2022-12-14 r83463)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 22.04.1 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## time zone: UTC
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] patchwork_1.1.2 ggplot2_3.4.0 echoplot_0.99.6 BiocStyle_2.27.0
##
## loaded via a namespace (and not attached):
## [1] fs_1.5.2 ProtGenerics_1.31.0
## [3] matrixStats_0.63.0 bitops_1.0-7
## [5] EnsDb.Hsapiens.v75_2.99.0 httr_1.4.4
## [7] RColorBrewer_1.1-3 Rgraphviz_2.43.0
## [9] tools_4.3.0 backports_1.4.1
## [11] utf8_1.2.2 R6_2.5.1
## [13] DT_0.26 lazyeval_0.2.2
## [15] withr_2.5.0 prettyunits_1.1.1
## [17] GGally_2.1.2 gridExtra_2.3
## [19] cli_3.5.0 Biobase_2.59.0
## [21] textshaping_0.3.6 labeling_0.4.2
## [23] ggbio_1.47.0 sass_0.4.4
## [25] mvtnorm_1.1-3 readr_2.1.3
## [27] proxy_0.4-27 pkgdown_2.0.7
## [29] Rsamtools_2.15.0 systemfonts_1.0.4
## [31] foreign_0.8-84 R.utils_2.12.2
## [33] dichromat_2.0-0.1 BSgenome_1.67.1
## [35] maps_3.4.1 readxl_1.4.1
## [37] rstudioapi_0.14 RSQLite_2.2.20
## [39] httpcode_0.3.0 pals_1.7
## [41] generics_0.1.3 BiocIO_1.9.1
## [43] echoconda_0.99.9 dplyr_1.0.10
## [45] zip_2.2.2 Matrix_1.5-3
## [47] interp_1.1-3 fansi_1.0.3
## [49] DescTools_0.99.47 S4Vectors_0.37.3
## [51] R.methodsS3_1.8.2 lifecycle_1.0.3
## [53] yaml_2.3.6 SummarizedExperiment_1.29.1
## [55] BiocFileCache_2.7.1 grid_4.3.0
## [57] blob_1.2.3 crayon_1.5.2
## [59] dir.expiry_1.7.0 lattice_0.20-45
## [61] GenomicFeatures_1.51.2 KEGGREST_1.39.0
## [63] mapproj_1.2.9 pillar_1.8.1
## [65] knitr_1.41 GenomicRanges_1.51.4
## [67] rjson_0.2.21 osfr_0.2.9
## [69] boot_1.3-28.1 gld_2.6.6
## [71] codetools_0.2-18 glue_1.6.2
## [73] data.table_1.14.6 vctrs_0.5.1
## [75] png_0.1-8 XGR_1.1.8
## [77] cellranger_1.1.0 gtable_0.3.1
## [79] assertthat_0.2.1 cachem_1.0.6
## [81] dnet_1.1.7 xfun_0.36
## [83] openxlsx_4.2.5.1 survival_3.4-0
## [85] ellipsis_0.3.2 nlme_3.1-161
## [87] bit64_4.0.5 progress_1.2.2
## [89] filelock_1.0.2 GenomeInfoDb_1.35.8
## [91] rprojroot_2.0.3 bslib_0.4.2
## [93] rpart_4.1.19 colorspace_2.0-3
## [95] BiocGenerics_0.45.0 DBI_1.1.3
## [97] Hmisc_4.7-2 nnet_7.3-18
## [99] Exact_3.2 tidyselect_1.2.0
## [101] bit_4.0.5 compiler_4.3.0
## [103] curl_4.3.3 graph_1.77.1
## [105] htmlTable_2.4.1 expm_0.999-6
## [107] basilisk.utils_1.11.1 xml2_1.3.3
## [109] desc_1.4.2 DelayedArray_0.25.0
## [111] bookdown_0.31 rtracklayer_1.59.0
## [113] checkmate_2.1.0 scales_1.2.1
## [115] hexbin_1.28.2 RBGL_1.75.0
## [117] echoLD_0.99.9 RCircos_1.2.2
## [119] rappdirs_0.3.3 stringr_1.5.0
## [121] supraHex_1.37.0 digest_0.6.31
## [123] piggyback_0.1.4 rmarkdown_2.19
## [125] basilisk_1.11.2 XVector_0.39.0
## [127] htmltools_0.5.4 pkgconfig_2.0.3
## [129] jpeg_0.1-10 base64enc_0.1-3
## [131] MatrixGenerics_1.11.0 echodata_0.99.16
## [133] highr_0.10 dbplyr_2.2.1
## [135] fastmap_1.1.0 ensembldb_2.23.1
## [137] rlang_1.0.6 htmlwidgets_1.6.0
## [139] farver_2.1.1 jquerylib_0.1.4
## [141] jsonlite_1.8.4 BiocParallel_1.33.7
## [143] R.oo_1.25.0 VariantAnnotation_1.45.0
## [145] RCurl_1.98-1.9 magrittr_2.0.3
## [147] Formula_1.2-4 GenomeInfoDbData_1.2.9
## [149] ggnetwork_0.5.10 munsell_0.5.0
## [151] Rcpp_1.0.9 ape_5.6-2
## [153] ggnewscale_0.4.8 reticulate_1.26
## [155] stringi_1.7.8 rootSolve_1.8.2.3
## [157] zlibbioc_1.45.0 MASS_7.3-58.1
## [159] plyr_1.8.8 parallel_4.3.0
## [161] ggrepel_0.9.2 snpStats_1.49.0
## [163] lmom_2.9 deldir_1.0-6
## [165] echoannot_0.99.10 Biostrings_2.67.0
## [167] splines_4.3.0 hms_1.1.2
## [169] igraph_1.3.5 reshape2_1.4.4
## [171] biomaRt_2.55.0 stats4_4.3.0
## [173] crul_1.3 XML_3.99-0.13
## [175] evaluate_0.19 latticeExtra_0.6-30
## [177] biovizBase_1.47.0 BiocManager_1.30.19
## [179] tzdb_0.3.0 tidyr_1.2.1
## [181] purrr_1.0.0 reshape_0.8.9
## [183] echotabix_0.99.9 restfulr_0.0.15
## [185] AnnotationFilter_1.23.0 e1071_1.7-12
## [187] downloadR_0.99.6 class_7.3-20.1
## [189] ragg_1.2.4 OrganismDbi_1.41.0
## [191] tibble_3.1.8 memoise_2.0.1
## [193] AnnotationDbi_1.61.0 GenomicAlignments_1.35.0
## [195] IRanges_2.33.0 cluster_2.1.4