Unlike finemap_loci, you don't need to provide a topSNPs
data.frame. Instead, just manually provide the coordinates of the locus
you want to fine-map.
finemap_locus(
locus,
fullSS_path,
fullSS_genome_build = NULL,
results_dir = file.path(tempdir(), "results"),
dataset_name = "dataset_name",
dataset_type = "GWAS",
case_control = TRUE,
topSNPs = "auto",
force_new_subset = FALSE,
force_new_LD = FALSE,
force_new_finemap = FALSE,
finemap_methods = c("ABF", "FINEMAP", "SUSIE"),
finemap_args = NULL,
n_causal = 5,
credset_thresh = 0.95,
consensus_thresh = 2,
fillNA = 0,
conditioned_snps = NULL,
priors_col = NULL,
munged = FALSE,
colmap = echodata::construct_colmap(munged = munged),
compute_n = "ldsc",
LD_reference = "1KGphase3",
LD_genome_build = "hg19",
leadSNP_LD_block = FALSE,
superpopulation = "EUR",
download_method = "axel",
bp_distance = 5e+05,
min_POS = NA,
max_POS = NA,
min_MAF = NA,
trim_gene_limits = FALSE,
max_snps = NULL,
min_r2 = 0,
remove_variants = FALSE,
remove_correlates = FALSE,
query_by = "tabix",
qtl_suffixes = NULL,
plot_types = c("simple"),
zoom = "1x",
show_plot = TRUE,
tx_biotypes = NULL,
nott_epigenome = FALSE,
nott_show_placseq = FALSE,
nott_binwidth = 200,
nott_bigwig_dir = NULL,
xgr_libnames = NULL,
roadmap = FALSE,
roadmap_query = NULL,
remove_tmps = TRUE,
seed = 2022,
conda_env = "echoR_mini",
nThread = 1,
verbose = TRUE,
top_SNPs = deprecated(),
PP_threshold = deprecated(),
consensus_threshold = deprecated(),
plot.Nott_epigenome = deprecated(),
plot.Nott_show_placseq = deprecated(),
plot.Nott_binwidth = deprecated(),
plot.Nott_bigwig_dir = deprecated(),
plot.Roadmap = deprecated(),
plot.Roadmap_query = deprecated(),
plot.XGR_libnames = deprecated(),
server = deprecated(),
plot.types = deprecated(),
plot.zoom = deprecated(),
QTL_prefixes = deprecated(),
vcf_folder = deprecated(),
probe_path = deprecated(),
file_sep = deprecated,
chrom_col = deprecated(),
chrom_type = deprecated(),
position_col = deprecated(),
snp_col = deprecated(),
pval_col = deprecated(),
effect_col = deprecated(),
stderr_col = deprecated(),
tstat_col = deprecated(),
locus_col = deprecated(),
freq_col = deprecated(),
MAF_col = deprecated(),
A1_col = deprecated(),
A2_col = deprecated(),
gene_col = deprecated(),
N_cases_col = deprecated(),
N_controls_col = deprecated(),
N_cases = deprecated(),
N_controls = deprecated(),
proportion_cases = deprecated(),
sample_size = deprecated(),
PAINTOR_QTL_datasets = deprecated()
)Arguments
- locus
Locus name to fine-map (e.g.
"BIN1"). Can be named to indicate a specific gene within a QTL locus (e.g.c(ENSG00000136731="BIN1")).- fullSS_path
Path to the full summary statistics file (GWAS or QTL) that you want to fine-map. It is usually best to provide the absolute path rather than the relative path.
- fullSS_genome_build
Genome build of the full summary statistics (
fullSS_path). Can be "GRCH37" or "GRCH38" or one of their synonyms.. IffullSS_genome_build==NULLandmunged=TRUE, infers genome build (hg19 vs. hg38) from summary statistics using get_genome_builds.- results_dir
Where to store all results. IMPORTANT!: It is usually best to provide the absolute path rather than the relative path. This is especially important for FINEMAP.
- dataset_name
The name you want to assign to the dataset being fine-mapped, This will be used to name the subdirectory where your results will be stored (e.g. Data/GWAS/<dataset_name>). Don't use special characters (e.g.".", "/").
- dataset_type
The kind dataset you're fine-mapping (e.g. GWAS, eQTL, tQTL). This will also be used when creating the subdirectory where your results will be stored (e.g. Data/<dataset_type>/Kunkle_2019).
- case_control
Whether the summary statistics come from a case-control study (e.g. a GWAS of having Alzheimer's Disease or not) (
TRUE) or a quantitative study (e.g. a GWAS of height, or an eQTL) (FALSE).- topSNPs
A data.frame with the genomic coordinates of the lead SNP for each locus. The lead SNP will be used as the center of the window when extracting subset from the full GWAS/QTL summary statistics file. Only one SNP per Locus should be included. At minimum,
topSNPsshould include the following columns:- Locus
A unique name for each locus. Often, loci are named after a relevant gene (e.g. LRRK2) or based on the name/coordinates of the lead SNP (e.g. locus_chr12_40734202)
- CHR
The chromosome that the SNP is on. Can be "chr12" or "12" format.
- POS
The genomic position of the SNP (in basepairs)
- force_new_subset
By default, if a subset of the full summary stats file for a given locus is already present, then echolocatoR will just use the pre-existing file. Set
force_new_subset=Tto override this and extract a new subset. Subsets are saved in the following path structure: Data/\<dataset_type\>/\<dataset_name\>/\<locus\>/Multi-finemap/ \<locus\>_\<dataset_name\>_Multi-finemap.tsv.gz- force_new_LD
Force new LD subset.
- force_new_finemap
By default, if an fine-mapping results file for a given locus is already present, then echolocatoR will just use the preexisting file. Set
force_new_finemap=Tto override this and re-run fine-mapping.- finemap_methods
Which fine-mapping methods you want to use.
- finemap_args
A named nested list containing additional arguments for each fine-mapping method. e.g.
finemap_args = list(FINEMAP=list(), PAINTOR=list(method=""))- n_causal
The maximum number of potential causal SNPs per locus. This parameter is used somewhat differently by different fine-mapping tools. See tool-specific functions for details.
- credset_thresh
The minimum fine-mapped posterior probability for a SNP to be considered part of a Credible Set. For example,
credset_thresh=.95means that all Credible Set SNPs will be 95% Credible Set SNPs.- consensus_thresh
The minimum number of fine-mapping tools in which a SNP is in the Credible Set in order to be included in the "Consensus_SNP" column.
- fillNA
Value to fill LD matrix NAs with.
- conditioned_snps
Which SNPs to conditions on when fine-mapping with (e.g. COJO).
- priors_col
[Optional] Name of the a column in
datto extract SNP-wise prior probabilities from.- munged
Whether
fullSS_pathhave already been standardised/filtered full summary stats with format_sumstats. Ifmunged=FALSEyou'll need to provide the necessary column names to thecolmapargument.- colmap
Column name mappings in in
fullSS_path. Must be a named list. Can use construct_colmap to assist with this. This function can be used in two different ways:munged=FALSE: Whenmunged=FALSE, you will need to provide the necessary column names to thecolmapargument (default).munged=TRUE: Alternatively, instead of filling out each argument in construct_colmap, you can simply setmunged=TRUEiffullSS_pathhas already been munged with format_sumstats.
- compute_n
How to compute per-SNP sample size (new column "N").
If the column "N" is already present indat, this column will be used to extract per-SNP sample sizes and the argumentcompute_nwill be ignored.
If the column "N" is not present indat, one of the following options can be supplied tocompute_n:0: N will not be computed.>0: If any number >0 is provided, that value will be set as N for every row. **Note**: Computing N this way is incorrect and should be avoided if at all possible."sum": N will be computed as: cases (N_CAS) + controls (N_CON), so long as both columns are present."ldsc": N will be computed as effective sample size: Neff =(N_CAS+N_CON)*(N_CAS/(N_CAS+N_CON)) / mean((N_CAS/(N_CAS+N_CON))(N_CAS+N_CON)==max(N_CAS+N_CON))."giant": N will be computed as effective sample size: Neff = 2 / (1/N_CAS + 1/N_CON)."metal": N will be computed as effective sample size: Neff = 4 / (1/N_CAS + 1/N_CON).
- LD_reference
LD reference to use:
"1KGphase1" : 1000 Genomes Project Phase 1 (genome build: hg19).
"1KGphase3" : 1000 Genomes Project Phase 3 (genome build: hg19).
"UKB" : Pre-computed LD from a British European-decent subset of UK Biobank. Genome build : hg19
"<vcf_path>" : User-supplied path to a custom VCF file to compute LD matrix from.
Accepted formats: .vcf / .vcf.gz / .vcf.bgz
Genome build : defined by user withtarget_genome."<matrix_path>" : User-supplied path to a pre-computed LD matrix Accepted formats: .rds / .rda / .csv / .tsv / .txt
Genome build : defined by user withtarget_genome.
- LD_genome_build
Genome build of the LD panel. This is automatically assigned to the correct genome build for each LD panel except when the user supplies custom vcf/LD files.
- leadSNP_LD_block
Only return SNPs within the same LD block as the lead SNP (the SNP with the smallest p-value).
- superpopulation
Superpopulation to subset LD panel by (used only if
LD_referenceis "1KGphase1" or "1KGphase3"). See popDat_1KGphase1 and popDat_1KGphase3 for full tables of their respective samples.- download_method
"axel": Multi-threaded"wget": Single-threaded"download.file": Single-threaded"internal": Single-threaded (passed to download.file)"wininet": Single-threaded (passed to download.file)"libcurl": Single-threaded (passed to download.file)"curl": Single-threaded (passed to download.file)
- bp_distance
Distance around the lead SNP to include.
- min_POS
Minimum genomic position to include.
- max_POS
Maximum genomic position to include.
- min_MAF
Minimum Minor Allele Frequency (MAF) of SNPs to include.
- trim_gene_limits
If a gene name is supplied to this argument (e.g.
trim_gene_limits="BST"), only SNPs within the gene body will be included.- max_snps
Maximum number of SNPs to include.
- min_r2
Correlation threshold for
remove_correlates.- remove_variants
A list of SNP RSIDs to remove.
- remove_correlates
A list of SNPs. If provided, all SNPs that correlates with these SNPs (at r2>=
min_r2) will be removed from bothdatandLDlist items..- query_by
Choose which method you want to use to extract locus subsets from the full summary stats file. Methods include:
- "tabix"
Convert the full summary stats file in an indexed tabix file. Makes querying lightning fast after the initial conversion is done. (default)
- "coordinates"
Extract locus subsets using min/max genomic coordinates with awk.
- qtl_suffixes
If columns with QTL data is included in
dat, you can indicate which columns those are with one or more string suffixes (e.g.qtl_suffixes=c(".eQTL1",".eQTL2")to use the columns "P.QTL1", "Effect.QTL1", "P.QTL2", "Effect.QTL2").- plot_types
Which kinds of plots to include. Options:
"simple"Just plot the following tracks: GWAS, fine-mapping, gene models
"fancy"Additionally plot XGR annotation tracks (XGR, Roadmap, Nott2019). '
"LD"LD heatmap showing the 10 SNPs surrounding the lead SNP.
- zoom
Zoom into the center of the locus when plotting (without editing the fine-mapping results file). You can provide either:
The size of your plot window in terms of basepairs (e.g.
zoom=50000for a 50kb window).How much you want to zoom in (e.g.
zoom="1x"for the full locus,zoom="2x"for 2x zoom into the center of the locus, etc.).
You can pass a list of window sizes (e.g.
c(50000,100000,500000)) to automatically generate multiple views of each locus. This can even be a mix of different style inputs: e.g.c("1x","4.5x",25000).- show_plot
Print plot to screen.
- tx_biotypes
Transcript biotypes to include in the gene model track. By default (
NULL), all transcript biotypes will be included. See get_tx_biotypes for a full list of all available biotypes- nott_epigenome
Include tracks showing brain cell-type-specific epigenomic data from Nott et al. (2019).
- nott_show_placseq
Include track generated by NOTT2019_plac_seq_plot.
- nott_binwidth
When including Nott et al. (2019) epigenomic data in the track plots, adjust the bin width of the histograms.
- nott_bigwig_dir
Instead of pulling Nott et al. (2019) epigenomic data from the UCSC Genome Browser, use a set of local bigwig files.
- xgr_libnames
Passed to XGR_plot. Which XGR annotations to check overlap with. For full list of libraries see here. Passed to the
RData.customisedargument in xRDataLoader. Examples:"ENCODE_TFBS_ClusteredV3_CellTypes"
"ENCODE_DNaseI_ClusteredV3_CellTypes"
"Broad_Histone"
- roadmap
Find and plot annotations from Roadmap.
- roadmap_query
Only plot annotations from Roadmap whose metadata contains a string or any items from a list of strings (e.g.
"brain"orc("brain","liver","monocytes")).- remove_tmps
Whether to remove any temporary files (e.g. FINEMAP output files) after the pipeline is done running.
- seed
Set the seed for all functions where this is possible.
- conda_env
Conda environment to use.
- nThread
Number of threads to parallelise saving across.
- verbose
Print messages.
- top_SNPs
[deprecated]
- PP_threshold
[deprecated]
- consensus_threshold
[deprecated]
- plot.Nott_epigenome
[deprecated]
- plot.Nott_show_placseq
[deprecated]
- plot.Nott_binwidth
[deprecated]
- plot.Nott_bigwig_dir
[deprecated]
- plot.Roadmap
[deprecated]
- plot.Roadmap_query
[deprecated]
- plot.XGR_libnames
[deprecated]
- server
[deprecated]
- plot.types
[deprecated]
- plot.zoom
[deprecated]
- QTL_prefixes
[deprecated]
- vcf_folder
[deprecated]
- probe_path
[deprecated]
- file_sep
[deprecated]
- chrom_col
[deprecated]
- chrom_type
[deprecated]
- position_col
[deprecated]
- snp_col
[deprecated]
- pval_col
[deprecated]
- effect_col
[deprecated]
- stderr_col
[deprecated]
- tstat_col
[deprecated]
- locus_col
[deprecated]
- freq_col
[deprecated]
- MAF_col
[deprecated]
- A1_col
[deprecated]
- A2_col
[deprecated]
- gene_col
[deprecated]
- N_cases_col
[deprecated]
- N_controls_col
[deprecated]
- N_cases
[deprecated]
- N_controls
[deprecated]
- proportion_cases
[deprecated]
- sample_size
[deprecated]
- PAINTOR_QTL_datasets
[deprecated]
Details
The primary functions of echolocatoR that expedite fine-mapping by wrapping many other echolocatoR functions into one. Encompasses steps including:
- Subset & standardize
Extract subsets of the full summary stats GWAS or QTL file and reformat them to be compatible with echolocatoR's various functions
- Calculate linkage disequilibrium
Download and prepare the necessary LD matrix.
- Fine-map
Run various fine-mapping tools and merge the results into a single multi-finemap data.frame.
- Plot
Summarise the results in a multi-track plot for each locus.
See also
Other MAIN:
finemap_loci()
Examples
topSNPs <- echodata::topSNPs_Nalls2019
fullSS_path <- echodata::example_fullSS(dataset = "Nalls2019")
#> Writing file to ==> /tmp/Rtmp0dbVNr/nalls2019.fullSS_subset.tsv
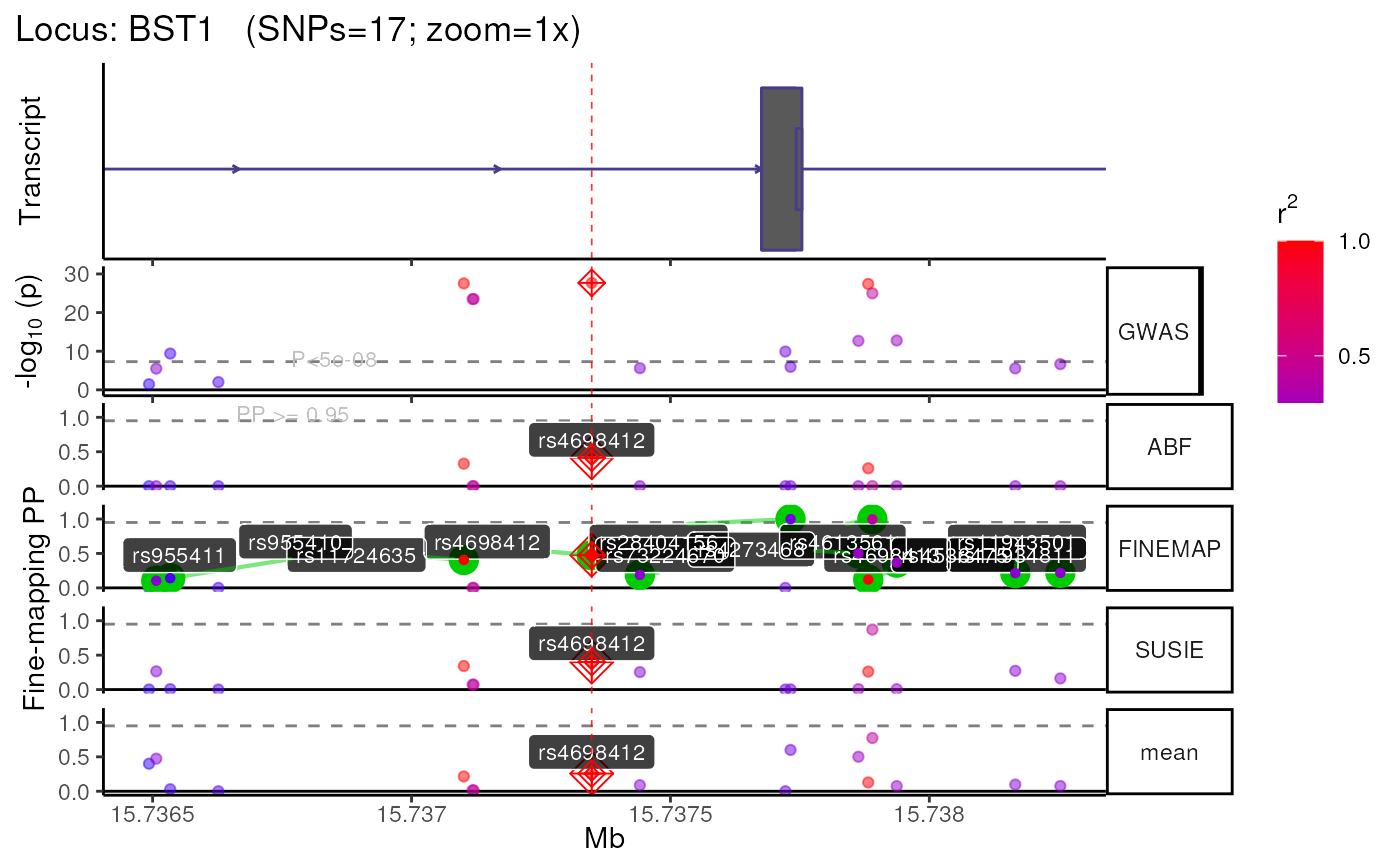
res <- echolocatoR::finemap_locus(
fullSS_path = fullSS_path,
topSNPs = topSNPs,
locus = "BST1",
finemap_methods = c("ABF","FINEMAP","SUSIE"),
dataset_name = "Nalls23andMe_2019",
fullSS_genome_build = "hg19",
bp_distance = 1000,
munged = TRUE)
#>
#> ────────────────────────────────────────────────────────────────────────────────
#>
#> ── Step 1 ▶▶▶ Query 🔎 ─────────────────────────────────────────────────────────
#>
#> ────────────────────────────────────────────────────────────────────────────────
#> + Importing pre-existing file: /tmp/Rtmp0dbVNr/results/GWAS/Nalls23andMe_2019/BST1/Multi-finemap/1KGphase3_LD.Multi-finemap.tsv.gz
#> Subset file looks good.
#>
#> ────────────────────────────────────────────────────────────────────────────────
#>
#> ── Step 2 ▶▶▶ Extract Linkage Disequilibrium 🔗 ────────────────────────────────
#>
#> ────────────────────────────────────────────────────────────────────────────────
#> LD_reference identified as: 1kg.
#> Previously computed LD_matrix detected. Importing: /tmp/Rtmp0dbVNr/results/GWAS/Nalls23andMe_2019/BST1/LD/BST1.1KGphase3_LD.RDS
#> LD_reference identified as: r.
#> Converting obj to sparseMatrix.
#> + FILTER:: Filtering by LD features.
#>
#> ────────────────────────────────────────────────────────────────────────────────
#>
#> ── Step 3 ▶▶▶ Filter SNPs 🚰 ───────────────────────────────────────────────────
#>
#> ────────────────────────────────────────────────────────────────────────────────
#> FILTER:: Filtering by SNP features.
#> + FILTER:: Post-filtered data: 17 x 29
#> + Subsetting LD matrix and dat to common SNPs...
#> Removing unnamed rows/cols
#> Replacing NAs with 0
#> + LD_matrix = 17 SNPs.
#> + dat = 17 SNPs.
#> + 17 SNPs in common.
#> Converting obj to sparseMatrix.
#>
#> ────────────────────────────────────────────────────────────────────────────────
#>
#> ── Step 4 ▶▶▶ Fine-map 🔊 ──────────────────────────────────────────────────────
#>
#> ────────────────────────────────────────────────────────────────────────────────
#> Gathering method sources.
#> Gathering method citations.
#> ++ Previously multi-finemapped results identified. Importing: /tmp/Rtmp0dbVNr/results/GWAS/Nalls23andMe_2019/BST1/Multi-finemap/1KGphase3_LD.Multi-finemap.tsv.gz
#> + Fine-mapping with 'ABF, FINEMAP, SUSIE' completed:
#>
#> ────────────────────────────────────────────────────────────────────────────────
#>
#> ── Step 5 ▶▶▶ Plot 📈 ──────────────────────────────────────────────────────────
#>
#> ────────────────────────────────────────────────────────────────────────────────
#> +-------- Locus Plot: BST1 --------+
#> + support_thresh = 2
#> + Calculating mean Posterior Probability (mean.PP)...
#> + 3 fine-mapping methods used.
#> + 12 Credible Set SNPs identified.
#> + 0 Consensus SNPs identified.
#> + Filling NAs in CS cols with 0.
#> + Filling NAs in PP cols with 0.
#> LD_matrix detected. Coloring SNPs by LD with lead SNP.
#> ++ echoplot:: GWAS full window track
#> ++ echoplot:: GWAS track
#> ++ echoplot:: Merged fine-mapping track
#> Melting PP and CS from 4 fine-mapping methods.
#> + echoplot:: Constructing SNP labels.
#> Adding SNP group labels to locus plot.
#> ++ echoplot:: Adding Gene model track.
#> Converting dat to GRanges object.
#> max_transcripts= 1 .
#> 1 transcripts from 1 genes returned.
#> Fetching data...
#> OK
#> Parsing exons...
#> OK
#> Defining introns...
#> OK
#> Defining UTRs...
#> OK
#> Defining CDS...
#> OK
#> aggregating...
#> Done
#> Constructing graphics...
#> + Adding vertical lines to highlight SNP groups.
#> +>+>+>+>+ zoom = 1x +<+<+<+<+
#> + echoplot:: Get window suffix...
#> + echoplot:: Removing GWAS full window track @ zoom=1x
#> + Removing subplot margins...
#> + Reordering tracks...
#> + Ensuring last track shows genomic units.
#> + Aligning xlimits for each subplot...
#> + Checking track heights...
#> + echoplot:: Saving plot ==> /tmp/Rtmp0dbVNr/results/GWAS/Nalls23andMe_2019/BST1/multiview.BST1.1KGphase3.1x.png
#> Recording all `finemap_locus` arguments.