## Registered S3 method overwritten by 'GGally':
## method from
## +.gg ggplot2## ⠊⠉⠡⣀⣀⠊⠉⠡⣀⣀⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠
## ⠌⢁⡐⠉⣀⠊⢂⡐⠑⣀⠊⢂⡐⠑⣀⠊⢂⡐⠑⣀⠊⢂⡐⠑⣀⠊⢂⡐⠑⣀⠉⢂⡈⠑⣀⠉⢄⡈⠡⣀
## ⠌⡈⡐⢂⢁⠒⡈⡐⢂⢁⠒⡈⡐⢂⢁⠑⡈⡈⢄⢁⠡⠌⡈⠤⢁⠡⠌⡈⠤⢁⠡⠌⡈⡠⢁⢁⠊⡈⡐⢂## ## ── 🦇 🦇 🦇 e c h o l o c a t o R 🦇 🦇 🦇 ─────────────────────────────────## ## ── v2.0.3 ──────────────────────────────────────────────────────────────────────## ⠌⡈⡐⢂⢁⠒⡈⡐⢂⢁⠒⡈⡐⢂⢁⠑⡈⡈⢄⢁⠡⠌⡈⠤⢁⠡⠌⡈⠤⢁⠡⠌⡈⡠⢁⢁⠊⡈⡐⢂
## ⠌⢁⡐⠉⣀⠊⢂⡐⠑⣀⠊⢂⡐⠑⣀⠊⢂⡐⠑⣀⠊⢂⡐⠑⣀⠊⢂⡐⠑⣀⠉⢂⡈⠑⣀⠉⢄⡈⠡⣀
## ⠊⠉⠡⣀⣀⠊⠉⠡⣀⣀⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠
## ⓞ If you use echolocatoR or any of the echoverse subpackages, please cite:
## ▶ Brian M Schilder, Jack Humphrey, Towfique
## Raj (2021) echolocatoR: an automated
## end-to-end statistical and functional
## genomic fine-mapping pipeline,
## Bioinformatics; btab658,
## https://doi.org/10.1093/bioinformatics/btab658
## ⓞ Please report any bugs/feature requests on GitHub:
## ▶
## https://github.com/RajLabMSSM/echolocatoR/issues
## ⓞ Contributions are welcome!:
## ▶
## https://github.com/RajLabMSSM/echolocatoR/pulls## ## ────────────────────────────────────────────────────────────────────────────────QTL pipeline
- Here, we will use GWAS-eQTL colocalization results provided via the echolocatoR Fine-mapping Portal, from the publication:
Lopes, K.d.P., Snijders, G.J.L., Humphrey, J. et al. Genetic analysis of the human microglial transcriptome across brain regions, aging and disease pathologies. Nat Genet 54, 4–17 (2022). https://doi.org/10.1038/s41588-021-00976-y
Import QTL data
This data is actually merged GWAS-QTL colocalization results, but it contains all of the necessary columns from the original eQTL summary stats that we need to perform eQTL fine-mapping.
coloc_res <- echodata::get_Kunkle2019_coloc(return_path = TRUE)Prepare colmap
Prepare a column mapping object for the summary statistics. We’ll
reuse this for both the import_topSNPs and
finemap_loci steps.
colmap <- echodata::construct_colmap(
CHR = "chr",
POS = "pos",
N = "qtl.N",
SNP = "snp",
P = "qtl.pvalues",
Effect = "qtl.beta",
StdErr = "qtl.varbeta",
MAF = "qtl.MAF",
Locus = "Locus",
Gene = "gene")Prepare top_SNPs data.frame
- In this case, we don’t have a top SNPs file ready. So we’re just
going to make one directly from the full summary stats file itself
(NOTE: You can only use this approach if you can fit the entire
file in memory).
- In this case, you’ll want to make sure to set
grouping_vars=c("Locus","Gene")so that you get top SNPs for each eGene-locus pair (not just one SNP per locus).
topSNPs <- echodata::import_topSNPs(
topSS = coloc_res$path,
colmap = colmap,
## Important for QTLs: group by both Locus and Gene
grouping_vars = c("Locus","Gene"))## Loading required namespace: MungeSumstats## Renaming column: chr ==> CHR## Renaming column: pos ==> POS## Renaming column: snp ==> SNP## Renaming column: qtl.pvalues ==> P## Renaming column: qtl.beta ==> Effect## Renaming column: qtl.varbeta ==> StdErr## Renaming column: qtl.MAF ==> MAF## Renaming column: gene ==> Gene## Renaming column: qtl.N ==> N## [1] "+ Assigning Gene and Locus independently."## Standardising column headers.## First line of summary statistics file:## Locus SNP V.df1 z.df1 r.df1 lABF.df1 V.df2 z.df2 r.df2 lABF.df2 internal.sum.lABF SNP.PP.H4 gwas.pvalues gwas.beta gwas.varbeta gwas.MAF CHR POS A1 A2 gwas.N gwas.type s Gene P Effect StdErr MAF QTL_chr QTL_pos N qtl.type N_cases N_controls ## Returning unmapped column names without making them uppercase.## + Mapping colnames from MungeSumstats ==> echolocatoR
head(topSNPs)## Locus SNP V.df1 z.df1 r.df1 lABF.df1
## 1: ABCA7_ENSG00000160953 rs76951864 0.00227529 -1.2138365 0.9461792 -0.7639979
## 2: BIN1_ENSG00000136731 rs4663110 0.00021904 2.0405405 0.9945538 -0.5358563
## 3: CR1_ENSG00000266094 rs1518110 0.00030276 -1.0977011 0.9924879 -1.8476694
## 4: INPP5D_ENSG00000168918 rs1881492 0.00036100 0.1631579 0.9910557 -2.3451794
## 5: MS4A6A_ENSG00000149476 rs6591611 0.00028900 -1.4176471 0.9928268 -1.4710500
## 6: PILRA_ENSG00000106366 rs2227631 0.00020736 -0.1041667 0.9948427 -2.6282771
## V.df2 z.df2 r.df2 lABF.df2 internal.sum.lABF SNP.PP.H4
## 1: 0.007802613 -5.826752 0.2374864 3.8958871 3.1318892 6.231637e-08
## 2: 0.004149176 2.778507 0.4321435 1.3851521 0.8492958 1.013056e-25
## 3: 0.002913938 -3.976592 0.4592553 3.3237628 1.4760934 4.495411e-11
## 4: 0.012164546 -2.654756 0.1470877 0.4387679 -1.9064116 3.587352e-08
## 5: 0.015193274 2.675112 0.2112492 0.6372210 -0.8338290 8.201862e-13
## 6: 0.006858275 8.459513 0.5879401 20.5941901 17.9659130 9.907667e-01
## gwas.pvalues gwas.beta gwas.varbeta gwas.MAF CHR POS A1 A2 gwas.N
## 1: 0.22500 -0.0579 0.00227529 0.1054 19 374604 A G 94437
## 2: 0.04153 0.0302 0.00021904 0.6262 2 127922463 T C 94437
## 3: 0.27270 -0.0191 0.00030276 0.7803 1 206944861 A C 94437
## 4: 0.87100 0.0031 0.00036100 0.7763 2 233406998 T G 94437
## 5: 0.15760 -0.0241 0.00028900 0.2207 11 60405172 A C 94437
## 6: 0.91860 -0.0015 0.00020736 0.4056 7 100769538 A G 94437
## gwas.type s Gene P Effect StdErr Freq
## 1: cc 0.37 ENSG00000160953 8.31563e-09 -0.514691 0.007802613 0.08770
## 2: cc 0.37 ENSG00000136731 7.93573e-05 0.178975 0.004149176 0.34385
## 3: cc 0.37 ENSG00000266094 1.00516e-04 -0.214660 0.002913938 0.25363
## 4: cc 0.37 ENSG00000168918 6.42575e-06 -0.292801 0.012164546 0.14447
## 5: cc 0.37 ENSG00000149476 2.20256e-04 0.329737 0.015193274 0.18503
## 6: cc 0.37 ENSG00000106366 5.45534e-17 0.700572 0.006858275 0.42796
## QTL_chr QTL_pos N qtl.type N_cases N_controls
## 1: chr19 374604 90 quant 34941 59496
## 2: chr2 127164887 90 quant 34941 59496
## 3: chr1 206771516 90 quant 34941 59496
## 4: chr2 232542288 90 quant 34941 59496
## 5: chr11 60637699 90 quant 34941 59496
## 6: chr7 101126257 90 quant 34941 59496Run fine-mapping pipeline
res <- echolocatoR::finemap_loci(fullSS_path = coloc_res$path,
topSNPs = topSNPs,
## Let's just fine-map 1 locus for demo purposes
loci = topSNPs$Locus[1],
dataset_name = "Kunkle_2019.microgliaQTL",
dataset_type = "QTL",
bp_distance = 1000,
colmap = colmap,
show_plot = TRUE,
finemap_methods = c("ABF","FINEMAP","SUSIE") )## [1] "+ Assigning Gene and Locus independently."## Standardising column headers.## First line of summary statistics file:## Locus SNP V.df1 z.df1 r.df1 lABF.df1 V.df2 z.df2 r.df2 lABF.df2 internal.sum.lABF SNP.PP.H4 gwas.pvalues gwas.beta gwas.varbeta gwas.MAF CHR POS A1 A2 gwas.N gwas.type s Gene P Effect StdErr Freq QTL_chr QTL_pos N qtl.type N_cases N_controls ## Returning unmapped column names without making them uppercase.## + Mapping colnames from MungeSumstats ==> echolocatoR## WARNING:: fullSS_genome_build not provided. Assuming 'GRCH37'.## ┌─────────────────────────────────────────────────────────┐
## │ │
## │ )))> 🦇 ABCA7_ENSG00000160953 [locus 1 / 1] 🦇 <((( │
## │ │
## └─────────────────────────────────────────────────────────┘## ## ────────────────────────────────────────────────────────────────────────────────## ## ── Step 1 ▶▶▶ Query 🔎 ─────────────────────────────────────────────────────────## ## ────────────────────────────────────────────────────────────────────────────────## + Query Method: tabix## Constructing GRanges query using min/max ranges within a single chromosome.## query_dat is already a GRanges object. Returning directly.## ========= echotabix::convert =========## Converting full summary stats file to tabix format for fast querying.## Inferred format: 'table'## Explicit format: 'table'## Inferring comment_char from tabular header: 'Locus'## Determining chrom type from file header.## Chromosome format: 1## Detecting column delimiter.## Identified column separator: ,## WARNING: Columns must be tab-separated ('\t') in order to be sorted outside of R (which is more memory-efficient).
## Will instead import full data into R to sort and rewrite to disk.## Sorting rows by coordinates via data.table.## Constructing outputs## Using existing bgzipped file: /github/home/.cache/R/echodata/Microglia_all_regions_Kunkle_2019_COLOC.snp-level_select.tsv.bgz
## Set force_new=TRUE to override this.## Tabix-indexing file using: Rsamtools## Data successfully converted to bgzip-compressed, tabix-indexed format.## ========= echotabix::query =========## query_dat is already a GRanges object. Returning directly.## Inferred format: 'table'## Querying tabular tabix file using: Rsamtools.## Checking query chromosome style is correct.## Chromosome format: 1## Retrieving data.## Converting query results to data.table.## Processing query: 19:373604-375604## Adding 'query' column to results.## Retrieved data with 10 rows## Saving query ==> /tmp/RtmpVZRnW7/results/QTL/Kunkle_2019.microgliaQTL/ABCA7_ENSG00000160953/ABCA7_ENSG00000160953_Kunkle_2019.microgliaQTL_subset.tsv.gz## + Query: 10 SNPs x 34 columns.## Standardizing summary statistics subset.## Standardizing main column names.## ++ Preparing A1,A1 cols## ++ Preparing MAF,Freq cols.## ++ Could not infer MAF from Freq.## ++ Removing SNPs with MAF== 0 | NULL | NA or >1.## ++ Preparing N_cases,N_controls cols.## ++ Preparing proportion_cases col.## ++ Calculating proportion_cases from N_cases and N_controls.## Preparing sample size column (N).## Using existing 'N' column.## + Imputing t-statistic from Effect and StdErr.## + leadSNP missing. Assigning new one by min p-value.## ++ Ensuring Effect,StdErr,P are numeric.## ++ Ensuring 1 SNP per row and per genomic coordinate.## ++ Removing extra whitespace## + Standardized query: 10 SNPs x 37 columns.## ++ Saving standardized query ==> /tmp/RtmpVZRnW7/results/QTL/Kunkle_2019.microgliaQTL/ABCA7_ENSG00000160953/ABCA7_ENSG00000160953_Kunkle_2019.microgliaQTL_subset.tsv.gz## ## ────────────────────────────────────────────────────────────────────────────────## ## ── Step 2 ▶▶▶ Extract Linkage Disequilibrium 🔗 ────────────────────────────────## ## ────────────────────────────────────────────────────────────────────────────────## LD_reference identified as: 1kg.## Using 1000Genomes as LD reference panel.## Constructing GRanges query using min/max ranges across one or more chromosomes.## + as_blocks=TRUE: Will query a single range per chromosome that covers all regions requested (plus anything in between).## LD Reference Panel = 1KGphase3## Querying 1KG remote server.## Selecting 503 EUR individuals from 1kgphase3.## ========= echotabix::query =========## query_dat is already a GRanges object. Returning directly.## Explicit format: 'vcf'## Querying VCF tabix file.## Querying VCF file using: VariantAnnotation## Checking query chromosome style is correct.## Chromosome format: 1## Filtering query to 503 samples and returning ScanVcfParam object.## Retrieving data.## Time difference of 4 secs## Removing 10 / 99 non-overlapping SNPs.## Saving VCF subset ==> /tmp/RtmpVZRnW7/VCF/RtmpVZRnW7.chr19-373673-375039.ALL.chr19.phase3_shapeit2_mvncall_integrated_v5a.20130502.genotypes.vcf.bgz## Warning in .Call(.make_vcf_geno, filename, fixed, names(geno), as.list(geno), :
## converting NULL pointer to R NULL## Time difference of 0.4 secs## Retrieved data with 10 rows across 503 samples.## echoLD::snpStats:: `MAF` column already present.## echoLD:snpStats:: Computing pairwise LD between 10 SNPs across 503 individuals (stats = R).## Time difference of 0 secs## 10 x 10 LD_matrix (sparse)## Converting obj to sparseMatrix.## Saving sparse LD matrix ==> /tmp/RtmpVZRnW7/results/QTL/Kunkle_2019.microgliaQTL/ABCA7_ENSG00000160953/LD/ABCA7_ENSG00000160953.1KGphase3_LD.RDS## Removing 1 temp files.## + FILTER:: Filtering by LD features.## ## ────────────────────────────────────────────────────────────────────────────────## ## ── Step 3 ▶▶▶ Filter SNPs 🚰 ───────────────────────────────────────────────────## ## ────────────────────────────────────────────────────────────────────────────────## FILTER:: Filtering by SNP features.## + FILTER:: Post-filtered data: 10 x 37## + Subsetting LD matrix and dat to common SNPs...## Removing unnamed rows/cols## Replacing NAs with 0## + LD_matrix = 10 SNPs.## + dat = 10 SNPs.## + 10 SNPs in common.## Converting obj to sparseMatrix.## ## ────────────────────────────────────────────────────────────────────────────────## ## ── Step 4 ▶▶▶ Fine-map 🔊 ──────────────────────────────────────────────────────## ## ────────────────────────────────────────────────────────────────────────────────## Gathering method sources.## Gathering method citations.## Preparing sample size column (N).## Using existing 'N' column.## Gathering method sources.## Gathering method citations.## ABF
## ✅ All required columns present.
## FINEMAP
## ✅ All required columns present.
## ✅ All optional columns present.
## SUSIE
## ✅ All required columns present.
## ✅ All optional columns present.## ++ Fine-mapping using 3 tool(s): ABF, FINEMAP, SUSIE##
## +++ Multi-finemap:: ABF +++## Using all default values for finemap_args$ABF## Preparing sample size column (N).## Using existing 'N' column.## Running ABF.## ++ Credible Set SNPs identified = 1## ++ Merging ABF results with multi-finemap data.##
## +++ Multi-finemap:: FINEMAP +++## Using all default values for finemap_args$FINEMAP## Preparing sample size column (N).## Using existing 'N' column.## + Subsetting LD matrix and dat to common SNPs...## Removing unnamed rows/cols## Replacing NAs with 0## + LD_matrix = 10 SNPs.## + dat = 10 SNPs.## + 10 SNPs in common.## Converting obj to sparseMatrix.## Constructing master file.## Constructing data.z file.## Constructing data.ld file.## FINEMAP path: /github/home/.cache/R/echofinemap/FINEMAP/finemap_v1.4.1_x86_64/finemap_v1.4.1_x86_64## Inferred FINEMAP version: 1.4.1## Running FINEMAP.## cd .../ABCA7_ENSG00000160953 &&
## .../finemap_v1.4.1_x86_64
##
## --sss
##
## --in-files .../master
##
## --log
##
## --n-causal-snps 5
##
## |--------------------------------------|
## | Welcome to FINEMAP v1.4.1 |
## | |
## | (c) 2015-2022 University of Helsinki |
## | |
## | Help : |
## | - ./finemap --help |
## | - www.finemap.me |
## | - www.christianbenner.com |
## | |
## | Contact : |
## | - finemap@christianbenner.com |
## | - matti.pirinen@helsinki.fi |
## |--------------------------------------|
##
## --------
## SETTINGS
## --------
## - dataset : all
## - corr-config : 0.95
## - n-causal-snps : 5
## - n-configs-top : 50000
## - n-conv-sss : 100
## - n-iter : 100000
## - n-threads : 1
## - prior-k0 : 0
## - prior-std : 0.05
## - prob-conv-sss-tol : 0.001
## - prob-cred-set : 0.95
##
## ------------
## FINE-MAPPING (1/1)
## ------------
## - GWAS summary stats : FINEMAP/data.z
## - SNP correlations : FINEMAP/data.ld
## - Causal SNP stats : FINEMAP/data.snp
## - Causal configurations : FINEMAP/data.config
## - Credible sets : FINEMAP/data.cred
## - Log file : FINEMAP/data.log_sss
##
- Reading summary statistics : +
- Reading summary statistics : -
- Reading summary stats : done!
- Reading SNP correlations : 0.000%
- Reading SNP correlations : done!
- Reading input : done!
##
##
- Estimating residual variance : +
- Estimating residual variance : -
- Updated prior SD of effect sizes : 0.05 0.108 0.234 0.505
##
## - Number of GWAS samples : 90
## - Number of SNPs : 10
## - Prior-Pr(# of causal SNPs is k) :
## (0 -> 0)
## 1 -> 0.595
## 2 -> 0.297
## 3 -> 0.0881
## 4 -> 0.0171
## 5 -> 0.00229
## 0 configurations evaluated (0.000/100%)
- 10 configurations evaluated (0.000/100%)
- 10 configurations evaluated (0.000/100%)
- 10 configurations evaluated (0.050/100%)
- 10 configurations evaluated (0.100/100%) : converged after 100 iterations
##
- Computing causal SNP statistics : 10.000%
- Computing causal SNP statistics : done!
## - Regional SNP heritability : 0.922 (SD: 0.0298 ; 95% CI: [0.864,0.979])
## - Log10-BF of >= one causal SNP : 28.8
## - Post-expected # of causal SNPs : 1
## - Post-Pr(# of causal SNPs is k) :
## (0 -> 0)
## 1 -> 1
## 2 -> 0
## 3 -> 0
## 4 -> 0
## 5 -> 0
## - Computing credible sets :
- Writing causal configurations : 10.000%
- Writing causal SNP statistics : 10.000%
- Writing output : done!
## - Run time : 0 hours, 0 minutes, 0 seconds## 1 data.cred* file(s) found in the same subfolder.## Selected file based on postPr_k: data.cred1## Importing conditional probabilities (.cred file).## No configurations were causal at PP>=0.95.## Importing marginal probabilities (.snp file).## Importing configuration probabilities (.config file).## FINEMAP was unable to identify any credible sets at PP>=0.95.## ++ Credible Set SNPs identified = 0## ++ Merging FINEMAP results with multi-finemap data.##
## +++ Multi-finemap:: SUSIE +++## Using all default values for finemap_args$SUSIE## Loading required namespace: Rfast## Preparing sample size column (N).## Using existing 'N' column.## + SUSIE:: sample_size=90## + Subsetting LD matrix and dat to common SNPs...## Removing unnamed rows/cols## Replacing NAs with 0## + LD_matrix = 10 SNPs.## + dat = 10 SNPs.## + 10 SNPs in common.## Converting obj to sparseMatrix.## + SUSIE:: Using `susie_rss()` from susieR v0.12.27## + SUSIE:: Extracting Credible Sets.## ++ Credible Set SNPs identified = 5## ++ Merging SUSIE results with multi-finemap data.## Identifying Consensus SNPs...## + support_thresh = 2## + Calculating mean Posterior Probability (mean.PP)...## + 3 fine-mapping methods used.## + 6 Credible Set SNPs identified.## + 0 Consensus SNPs identified.## Saving merged finemapping results ==> /tmp/RtmpVZRnW7/results/QTL/Kunkle_2019.microgliaQTL/ABCA7_ENSG00000160953/Multi-finemap/1KGphase3_LD.Multi-finemap.tsv.gz## + Fine-mapping with 'ABF, FINEMAP, SUSIE' completed:## ## ────────────────────────────────────────────────────────────────────────────────## ## ── Step 5 ▶▶▶ Plot 📈 ──────────────────────────────────────────────────────────## ## ────────────────────────────────────────────────────────────────────────────────## +-------- Locus Plot: ABCA7_ENSG00000160953 --------+## + support_thresh = 2## + Calculating mean Posterior Probability (mean.PP)...## + 3 fine-mapping methods used.## + 6 Credible Set SNPs identified.## + 0 Consensus SNPs identified.## + Filling NAs in CS cols with 0.## + Filling NAs in PP cols with 0.## LD_matrix detected. Coloring SNPs by LD with lead SNP.## ++ echoplot:: QTL full window track## ++ echoplot:: QTL track## ++ echoplot:: Merged fine-mapping track## Melting PP and CS from 4 fine-mapping methods.## + echoplot:: Constructing SNP labels.## Adding SNP group labels to locus plot.## ++ echoplot:: Adding Gene model track.## Converting dat to GRanges object.## Loading required namespace: EnsDb.Hsapiens.v75## max_transcripts= 1 .## 1 transcripts from 1 genes returned.## Fetching data...OK
## Parsing exons...OK
## Defining introns...OK
## Defining UTRs...OK
## Defining CDS...OK
## aggregating...
## Done
## Constructing graphics...
## + Adding vertical lines to highlight SNP groups.
## +>+>+>+>+ zoom = 1x +<+<+<+<+
## + echoplot:: Get window suffix...
## + echoplot:: Removing QTL full window track @ zoom=1x
## + Removing subplot margins...
## + Reordering tracks...
## + Ensuring last track shows genomic units.
## + Aligning xlimits for each subplot...
## + Checking track heights...
## + echoplot:: Saving plot ==> /tmp/RtmpVZRnW7/results/QTL/Kunkle_2019.microgliaQTL/ABCA7_ENSG00000160953/multiview.ABCA7_ENSG00000160953.1KGphase3.1x.png
## Found more than one class "simpleUnit" in cache; using the first, from namespace 'hexbin'
## Also defined by 'ggbio'
## Recording all `finemap_locus` arguments.
## Formatting locus results.
##
## ────────────────────────────────────────────────────────────────────────────────
##
## ── Step 6 ▶▶▶ Postprocess data 🎁 ──────────────────────────────────────────────
##
## ────────────────────────────────────────────────────────────────────────────────
## Returning results as nested list.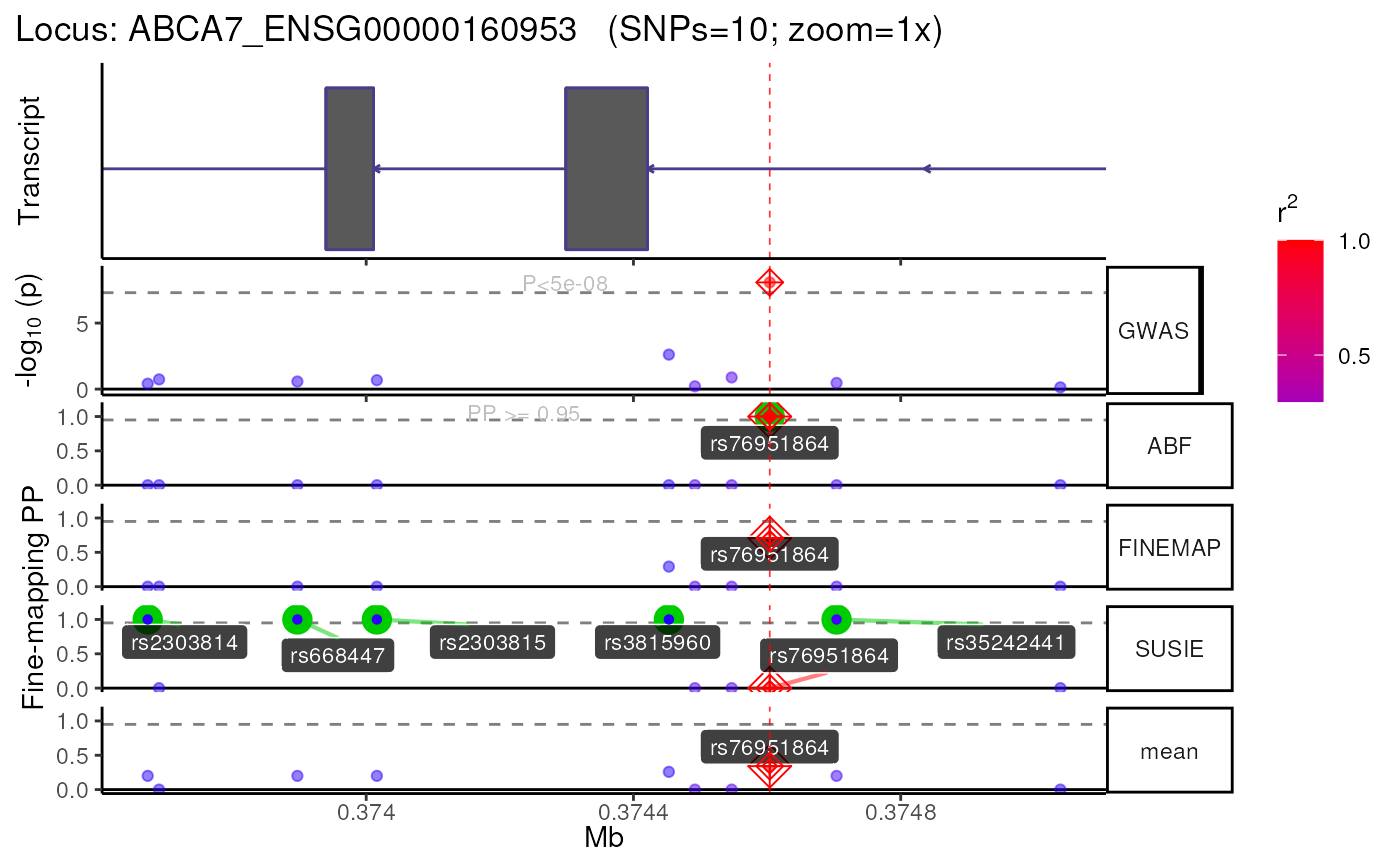
## All loci done in: 0.27 min
## Session info
utils::sessionInfo()## R Under development (unstable) (2023-01-11 r83598)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 22.04.1 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## time zone: UTC
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] snpStats_1.49.0 Matrix_1.5-3 survival_3.5-0 echolocatoR_2.0.3
## [5] BiocStyle_2.27.0
##
## loaded via a namespace (and not attached):
## [1] ProtGenerics_1.31.0 fs_1.5.2
## [3] matrixStats_0.63.0 bitops_1.0-7
## [5] EnsDb.Hsapiens.v75_2.99.0 httr_1.4.4
## [7] RColorBrewer_1.1-3 Rgraphviz_2.43.0
## [9] tools_4.3.0 backports_1.4.1
## [11] utf8_1.2.2 R6_2.5.1
## [13] DT_0.27 lazyeval_0.2.2
## [15] withr_2.5.0 prettyunits_1.1.1
## [17] GGally_2.1.2 gridExtra_2.3
## [19] cli_3.6.0 Biobase_2.59.0
## [21] textshaping_0.3.6 labeling_0.4.2
## [23] ggbio_1.47.0 sass_0.4.4
## [25] mvtnorm_1.1-3 readr_2.1.3
## [27] proxy_0.4-27 pkgdown_2.0.7
## [29] mixsqp_0.3-48 Rsamtools_2.15.1
## [31] systemfonts_1.0.4 foreign_0.8-84
## [33] R.utils_2.12.2 dichromat_2.0-0.1
## [35] maps_3.4.1 BSgenome_1.67.3
## [37] readxl_1.4.1 susieR_0.12.27
## [39] pals_1.7 rstudioapi_0.14
## [41] RSQLite_2.2.20 httpcode_0.3.0
## [43] generics_0.1.3 BiocIO_1.9.1
## [45] echoconda_0.99.9 dplyr_1.0.10
## [47] zip_2.2.2 interp_1.1-3
## [49] fansi_1.0.3 DescTools_0.99.47
## [51] S4Vectors_0.37.3 catalogueR_1.0.1
## [53] R.methodsS3_1.8.2 lifecycle_1.0.3
## [55] yaml_2.3.6 SummarizedExperiment_1.29.1
## [57] BiocFileCache_2.7.1 echoplot_0.99.6
## [59] grid_4.3.0 blob_1.2.3
## [61] crayon_1.5.2 dir.expiry_1.7.0
## [63] lattice_0.20-45 GenomicFeatures_1.51.2
## [65] mapproj_1.2.11 KEGGREST_1.39.0
## [67] MungeSumstats_1.7.15 pillar_1.8.1
## [69] knitr_1.41 GenomicRanges_1.51.4
## [71] rjson_0.2.21 osfr_0.2.9
## [73] boot_1.3-28.1 gld_2.6.6
## [75] codetools_0.2-18 glue_1.6.2
## [77] data.table_1.14.6 coloc_5.1.0.1
## [79] vctrs_0.5.1 png_0.1-8
## [81] XGR_1.1.8 cellranger_1.1.0
## [83] gtable_0.3.1 assertthat_0.2.1
## [85] cachem_1.0.6 dnet_1.1.7
## [87] xfun_0.36 openxlsx_4.2.5.1
## [89] Rfast_2.0.6 gargle_1.2.1
## [91] ellipsis_0.3.2 nlme_3.1-161
## [93] bit64_4.0.5 progress_1.2.2
## [95] filelock_1.0.2 googleAuthR_2.0.0
## [97] GenomeInfoDb_1.35.12 rprojroot_2.0.3
## [99] bslib_0.4.2 irlba_2.3.5.1
## [101] rpart_4.1.19 colorspace_2.0-3
## [103] BiocGenerics_0.45.0 DBI_1.1.3
## [105] Hmisc_4.7-2 nnet_7.3-18
## [107] Exact_3.2 tidyselect_1.2.0
## [109] bit_4.0.5 compiler_4.3.0
## [111] curl_5.0.0 graph_1.77.1
## [113] htmlTable_2.4.1 expm_0.999-7
## [115] basilisk.utils_1.11.1 xml2_1.3.3
## [117] desc_1.4.2 DelayedArray_0.25.0
## [119] bookdown_0.32 rtracklayer_1.59.1
## [121] checkmate_2.1.0 scales_1.2.1
## [123] hexbin_1.28.2 echoLD_0.99.9
## [125] RBGL_1.75.0 RCircos_1.2.2
## [127] rappdirs_0.3.3 stringr_1.5.0
## [129] supraHex_1.37.0 digest_0.6.31
## [131] piggyback_0.1.4 rmarkdown_2.20
## [133] basilisk_1.11.2 XVector_0.39.0
## [135] htmltools_0.5.4 pkgconfig_2.0.3
## [137] jpeg_0.1-10 base64enc_0.1-3
## [139] MatrixGenerics_1.11.0 echodata_0.99.16
## [141] highr_0.10 ensembldb_2.23.1
## [143] dbplyr_2.3.0 fastmap_1.1.0
## [145] rlang_1.0.6 htmlwidgets_1.6.1
## [147] farver_2.1.1 echofinemap_0.99.5
## [149] jquerylib_0.1.4 jsonlite_1.8.4
## [151] BiocParallel_1.33.9 R.oo_1.25.0
## [153] VariantAnnotation_1.45.0 RCurl_1.98-1.9
## [155] magrittr_2.0.3 Formula_1.2-4
## [157] GenomeInfoDbData_1.2.9 ggnetwork_0.5.10
## [159] patchwork_1.1.2 munsell_0.5.0
## [161] Rcpp_1.0.9 ggnewscale_0.4.8
## [163] ape_5.6-2 viridis_0.6.2
## [165] reticulate_1.27 RcppZiggurat_0.1.6
## [167] stringi_1.7.12 rootSolve_1.8.2.3
## [169] zlibbioc_1.45.0 MASS_7.3-58.1
## [171] plyr_1.8.8 parallel_4.3.0
## [173] ggrepel_0.9.2 lmom_2.9
## [175] deldir_1.0-6 echoannot_0.99.10
## [177] Biostrings_2.67.0 splines_4.3.0
## [179] hms_1.1.2 igraph_1.3.5
## [181] reshape2_1.4.4 biomaRt_2.55.0
## [183] stats4_4.3.0 crul_1.3
## [185] XML_3.99-0.13 evaluate_0.20
## [187] latticeExtra_0.6-30 biovizBase_1.47.0
## [189] BiocManager_1.30.19 tzdb_0.3.0
## [191] tidyr_1.2.1 purrr_1.0.1
## [193] reshape_0.8.9 ggplot2_3.4.0
## [195] echotabix_0.99.9 restfulr_0.0.15
## [197] AnnotationFilter_1.23.0 e1071_1.7-12
## [199] downloadR_0.99.6 viridisLite_0.4.1
## [201] class_7.3-20.1 ragg_1.2.5
## [203] OrganismDbi_1.41.0 tibble_3.1.8
## [205] memoise_2.0.1 AnnotationDbi_1.61.0
## [207] GenomicAlignments_1.35.0 IRanges_2.33.0
## [209] cluster_2.1.4