Get started
¶ Author:
Brian M. Schilder ¶
¶ Updated:
Jan-20-2023 ¶
Source: vignettes/echolocatoR.Rmd
echolocatoR.Rmd## Registered S3 method overwritten by 'GGally':
## method from
## +.gg ggplot2## ⠊⠉⠡⣀⣀⠊⠉⠡⣀⣀⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠
## ⠌⢁⡐⠉⣀⠊⢂⡐⠑⣀⠊⢂⡐⠑⣀⠊⢂⡐⠑⣀⠊⢂⡐⠑⣀⠊⢂⡐⠑⣀⠉⢂⡈⠑⣀⠉⢄⡈⠡⣀
## ⠌⡈⡐⢂⢁⠒⡈⡐⢂⢁⠒⡈⡐⢂⢁⠑⡈⡈⢄⢁⠡⠌⡈⠤⢁⠡⠌⡈⠤⢁⠡⠌⡈⡠⢁⢁⠊⡈⡐⢂## ## ── 🦇 🦇 🦇 e c h o l o c a t o R 🦇 🦇 🦇 ─────────────────────────────────## ## ── v2.0.3 ──────────────────────────────────────────────────────────────────────## ⠌⡈⡐⢂⢁⠒⡈⡐⢂⢁⠒⡈⡐⢂⢁⠑⡈⡈⢄⢁⠡⠌⡈⠤⢁⠡⠌⡈⠤⢁⠡⠌⡈⡠⢁⢁⠊⡈⡐⢂
## ⠌⢁⡐⠉⣀⠊⢂⡐⠑⣀⠊⢂⡐⠑⣀⠊⢂⡐⠑⣀⠊⢂⡐⠑⣀⠊⢂⡐⠑⣀⠉⢂⡈⠑⣀⠉⢄⡈⠡⣀
## ⠊⠉⠡⣀⣀⠊⠉⠡⣀⣀⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠⠊⠉⠢⣀⡠
## ⓞ If you use echolocatoR or any of the echoverse subpackages, please cite:
## ▶ Brian M Schilder, Jack Humphrey, Towfique
## Raj (2021) echolocatoR: an automated
## end-to-end statistical and functional
## genomic fine-mapping pipeline,
## Bioinformatics; btab658,
## https://doi.org/10.1093/bioinformatics/btab658
## ⓞ Please report any bugs/feature requests on GitHub:
## ▶
## https://github.com/RajLabMSSM/echolocatoR/issues
## ⓞ Contributions are welcome!:
## ▶
## https://github.com/RajLabMSSM/echolocatoR/pulls## ## ────────────────────────────────────────────────────────────────────────────────Full pipeline
All examples below use data from the Parkinson’s disease GWAS by Nalls et al. (2019).
Prepare top_SNPs data.frame
- To enable rapid fine-mapping of many loci, you can create a
top_SNPsdata.frame
which contains the position of the lead/index SNP within each locus. -
finemap_loci()(see next step) will then use this info to extract subsets of the
full GWAS/QTL summary statistics using windows centered on each lead/index SNP. - The
topSSargument can either be a data.frame, or a path to a topSS file saved somehwere. Most common tabular data formats (e.g. .tsv, .csv, .xlsx) are accepted.
#### Load example top SNPs (pre-formatted) ####
topSS <- echodata::topSNPs_Nalls2019_raw
#### construct a column mapping object ####
colmap <- echodata::construct_colmap(P = "P, all studies",
Effect = "Beta, all studies",
Locus = "Nearest Gene",
Gene = "QTL Nominated Gene (nearest QTL)")
#### Import top SNPs ####
topSNPs <- echodata::import_topSNPs(
topSS = echodata::topSNPs_Nalls2019_raw,
colmap = colmap,
grouping_vars = "Locus Number")## Loading required namespace: MungeSumstats## Renaming column: P, all studies ==> P## Renaming column: Beta, all studies ==> Effect## Renaming column: Nearest Gene ==> Locus## Renaming column: QTL Nominated Gene (nearest QTL) ==> Gene## [1] "+ Assigning Gene and Locus independently."## Standardising column headers.## First line of summary statistics file:## SNP CHR BP Locus Gene Effect allele Other allele Effect allele frequency Effect SE, all studies P P, COJO, all studies P, random effects, all studies P, Conditional 23AndMe only P, 23AndMe only I2, all studies Freq1, previous studies Beta, previous studies StdErr, previous studies P, previous studies I2, previous studies Freq1, new studies Beta, new studies StdErr, new studies P, new studies I2, new studies Passes pooled 23andMe QC Known GWAS locus within 1MB Failed final filtering and QC Locus within 250KB Locus Number ## Returning unmapped column names without making them uppercase.## + Mapping colnames from MungeSumstats ==> echolocatoR
head(topSNPs)## SNP CHR POS Locus Gene A2 A1 Effect allele frequency
## 1: rs1941685 18 31304318 ASXL3 <NA> t g 0.4983
## 2: rs2280104 8 22525980 BIN3 BIN3 t c 0.3604
## 3: rs61169879 17 59917366 BRIP1 MED13 t c 0.1641
## 4: rs4698412 4 15737348 BST1 BST1 a g 0.5529
## 5: rs11950533 5 134199105 C5orf24 TXNDC15 a c 0.1020
## 6: rs9568188 13 49927732 CAB39L CAB39L t c 0.7397
## Effect SE, all studies P P, COJO, all studies
## 1: 0.0531 0.0094 1.69e-08 1.61e-08
## 2: 0.0556 0.0098 1.16e-08 1.40e-08
## 3: 0.0820 0.0134 9.28e-10 9.40e-10
## 4: 0.1035 0.0094 2.06e-28 9.73e-36
## 5: -0.0916 0.0158 7.16e-09 6.73e-09
## 6: 0.0617 0.0108 1.15e-08 1.11e-08
## P, random effects, all studies P, Conditional 23AndMe only P, 23AndMe only
## 1: 1.690e-08 1.64e-08 1.60e-08
## 2: 1.860e-06 4.35e-02 4.94e-02
## 3: 6.210e-06 9.07e-07 2.05e-06
## 4: 1.680e-19 1.05e-07 1.15e-07
## 5: 2.680e-08 5.08e-04 4.22e-04
## 6: 2.458e-04 4.29e-06 4.41e-06
## I2, all studies Freq1, previous studies Beta, previous studies
## 1: 0.0 0.4978 0.0507
## 2: 8.9 0.3595 0.0647
## 3: 16.4 0.1641 0.0849
## 4: 13.9 0.5547 0.1023
## 5: 1.9 0.1008 -0.0986
## 6: 21.4 0.7405 0.0657
## StdErr, previous studies P, previous studies I2, previous studies
## 1: 0.0113 6.82e-06 0.0
## 2: 0.0117 3.23e-08 15.0
## 3: 0.0163 1.90e-07 0.0
## 4: 0.0112 7.32e-20 49.3
## 5: 0.0190 2.05e-07 43.8
## 6: 0.0130 4.00e-07 39.8
## Freq1, new studies Beta, new studies StdErr, new studies P, new studies
## 1: 0.4995 0.0586 0.0171 6.147e-04
## 2: 0.3626 0.0350 0.0177 4.724e-02
## 3: 0.1641 0.0761 0.0236 1.242e-03
## 4: 0.5485 0.1062 0.0170 4.160e-10
## 5: 0.1048 -0.0756 0.0287 8.388e-03
## 6: 0.7380 0.0526 0.0196 7.353e-03
## I2, new studies Passes pooled 23andMe QC Known GWAS locus within 1MB
## 1: 14.3 T 0
## 2: 1.9 T 1
## 3: 23.7 T 0
## 4: 10.9 T 1
## 5: 0.0 T 0
## 6: 22.3 T 0
## Failed final filtering and QC Locus within 250KB Locus Number
## 1: 0 0 73
## 2: 0 0 39
## 3: 0 0 71
## 4: 0 0 20
## 5: 0 0 28
## 6: 0 0 53Path to full summary stats file
Since a full GWAS summary stats file would be too large to include within echolocatoR,
we instead provide an example subset of the full summary stats.To simulate how you’d actually use your own full summary stats file, we will save our example dataset to your computer (you can change the path to wherever you like).
We highly recommend munging your full summary stats using the Bioconductor package
MungeSumstatsfirst. It’s easy to use and very robust. It also means you don’t have to provide most column mapping arguments infinemap_lociwhenmunged=TRUE.
Here’s an example of how to munge your full summary stats file:
fullSS_path <- echodata::example_fullSS(munged = FALSE)
fullSS_path <- MungeSumstats::format_sumstats(path = fullSS_path, ref_genome = "GRCH37")We have already munged the following example summary stats for you.
fullSS_path <- echodata::example_fullSS(dataset = "Nalls2019")## Writing file to ==> /tmp/RtmpKaL0Dw/nalls2019.fullSS_subset.tsvRun fine-mapping pipeline
For a full description of all arguments, see
?finemap_loci.
Here are some key arguments:
-
results_dir: Where you want to store all of your
results.
-
finemap_methods: Which fine-mapping methods you want to
run. For a full list of currently supported methods, run the function
echofinemap::lfm().
-
bp_distance: Controls window size. Specifically,
bp_distanceis the number of basepairs upstream/downstream you want to extract for each locus. For example, if you want a 2Mb window (+/- 1Mb from the lead/index SNP intop_SNPs), setbp_distance=1e+06.
-
plot_zoom: Zoom in/out from the center of each locus when
producing the multiview plot.
You can adjust this separately frombp_distanceso that you don’t have rerun the whole pipeline each time (locus subsets, LD matrices, and fine-mapping results are all automatically saved in locus-specific folders).
Note: Please use the full absolute paths (instead of
relative paths) wherever possible (e.g. results_dir). This
is especially important for the tool FINEMAP.
results <- echolocatoR::finemap_loci(
fullSS_path = fullSS_path,
topSNPs = topSNPs,
loci = c("BST1","MEX3C"),
LD_reference = "1KGphase3",
dataset_name = "Nalls23andMe_2019",
fullSS_genome_build = "hg19",
bp_distance = 1000,
finemap_methods = c("ABF","SUSIE","FINEMAP"),
munged = TRUE)## [1] "+ Assigning Gene and Locus independently."## Standardising column headers.## First line of summary statistics file:## SNP CHR POS Locus Gene A2 A1 Effect allele frequency Effect SE, all studies P P, COJO, all studies P, random effects, all studies P, Conditional 23AndMe only P, 23AndMe only I2, all studies Freq1, previous studies Beta, previous studies StdErr, previous studies P, previous studies I2, previous studies Freq1, new studies Beta, new studies StdErr, new studies P, new studies I2, new studies Passes pooled 23andMe QC Known GWAS locus within 1MB Failed final filtering and QC Locus within 250KB Locus Number ## Returning unmapped column names without making them uppercase.## + Mapping colnames from MungeSumstats ==> echolocatoR## ┌────────────────────────────────────────┐
## │ │
## │ )))> 🦇 BST1 [locus 1 / 2] 🦇 <((( │
## │ │
## └────────────────────────────────────────┘## ## ────────────────────────────────────────────────────────────────────────────────## ## ── Step 1 ▶▶▶ Query 🔎 ─────────────────────────────────────────────────────────## ## ────────────────────────────────────────────────────────────────────────────────## + Query Method: tabix## Constructing GRanges query using min/max ranges within a single chromosome.## query_dat is already a GRanges object. Returning directly.## ========= echotabix::convert =========## Converting full summary stats file to tabix format for fast querying.## Inferred format: 'table'## Explicit format: 'table'## Inferring comment_char from tabular header: 'SNP'## Determining chrom type from file header.## Assuming fullSS_path summary stats have already been processed with MungeSumstats.## Chromosome format: 1## Detecting column delimiter.## Identified column separator: \t## Sorting rows by coordinates via bash.## Searching for header row with grep.## ( grep ^'SNP' .../nalls2019.fullSS_subset.tsv; grep
## -v ^'SNP' .../nalls2019.fullSS_subset.tsv | sort
## -k2,2n
## -k3,3n ) > .../file134aea4a0607_sorted.tsv## Constructing outputs## bgzipping file with Rsamtools.## Reading bgzipped file using: data.table## Header preview:## SNP CHR BP A1 A2 FREQ BETA SE P N_CAS N_CON
## 1: rs741214 4 13737637 C A 0.2341 0.0091 0.0119 0.4460 49053 1411006
## 2: rs79385021 4 13737659 T C 0.9109 0.0009 0.0198 0.9642 30435 974587
## 3: rs741215 4 13737767 C T 0.0733 0.0190 0.0194 0.3284 49053 1411006
## 4: rs759261 4 13737915 G A 0.4511 0.0075 0.0100 0.4543 49053 1411006
## 5: rs35006360 4 13738014 G A 0.1148 -0.0195 0.0160 0.2208 49053 1411006## Tabix-indexing file using: Rsamtools## Data successfully converted to bgzip-compressed, tabix-indexed format.## ========= echotabix::query =========## query_dat is already a GRanges object. Returning directly.## Inferred format: 'table'## Querying tabular tabix file using: Rsamtools.## Checking query chromosome style is correct.## Chromosome format: 1## Retrieving data.## Converting query results to data.table.## Processing query: 4:15736348-15738348## Adding 'query' column to results.## Retrieved data with 17 rows## Saving query ==> /tmp/RtmpKaL0Dw/results/GWAS/Nalls23andMe_2019/BST1/BST1_Nalls23andMe_2019_subset.tsv.gz## + Query: 17 SNPs x 11 columns.## Standardizing summary statistics subset.## Standardizing main column names.## ++ Preparing A1,A1 cols## ++ Preparing MAF,Freq cols.## ++ Could not infer MAF from Freq.## ++ Preparing N_cases,N_controls cols.## ++ Preparing proportion_cases col.## ++ Calculating proportion_cases from N_cases and N_controls.## Preparing sample size column (N).## Computing effective sample size using the LDSC method:
## Neff = (N_CAS+N_CON) * (N_CAS/(N_CAS+N_CON)) / mean((N_CAS/(N_CAS+N_CON))[(N_CAS+N_CON)==max(N_CAS+N_CON)]))## + Mapping colnames from MungeSumstats ==> echolocatoR## + Imputing t-statistic from Effect and StdErr.## + leadSNP missing. Assigning new one by min p-value.## ++ Ensuring Effect,StdErr,P are numeric.## ++ Ensuring 1 SNP per row and per genomic coordinate.## ++ Removing extra whitespace## + Standardized query: 17 SNPs x 15 columns.## ++ Saving standardized query ==> /tmp/RtmpKaL0Dw/results/GWAS/Nalls23andMe_2019/BST1/BST1_Nalls23andMe_2019_subset.tsv.gz## ## ────────────────────────────────────────────────────────────────────────────────## ## ── Step 2 ▶▶▶ Extract Linkage Disequilibrium 🔗 ────────────────────────────────## ## ────────────────────────────────────────────────────────────────────────────────## LD_reference identified as: 1kg.## Using 1000Genomes as LD reference panel.## Constructing GRanges query using min/max ranges across one or more chromosomes.## + as_blocks=TRUE: Will query a single range per chromosome that covers all regions requested (plus anything in between).## LD Reference Panel = 1KGphase3## Querying 1KG remote server.## Selecting 503 EUR individuals from 1kgphase3.## ========= echotabix::query =========## query_dat is already a GRanges object. Returning directly.## Explicit format: 'vcf'## Querying VCF tabix file.## Querying VCF file using: VariantAnnotation## Checking query chromosome style is correct.## Chromosome format: 1## Filtering query to 503 samples and returning ScanVcfParam object.## Retrieving data.## Time difference of 4.5 secs## Removing 17 / 66 non-overlapping SNPs.## Saving VCF subset ==> /tmp/RtmpKaL0Dw/VCF/RtmpKaL0Dw.chr4-15736493-15738253.ALL.chr4.phase3_shapeit2_mvncall_integrated_v5a.20130502.genotypes.vcf.bgz## Warning in .Call(.make_vcf_geno, filename, fixed, names(geno), as.list(geno), :
## converting NULL pointer to R NULL## Time difference of 0.4 secs## Retrieved data with 17 rows across 503 samples.## echoLD::snpStats:: Filling `MAF` column with MAF from LD panel.## echoLD:snpStats:: Computing pairwise LD between 17 SNPs across 503 individuals (stats = R).## Time difference of 0 secs## 17 x 17 LD_matrix (sparse)## Converting obj to sparseMatrix.## Saving sparse LD matrix ==> /tmp/RtmpKaL0Dw/results/GWAS/Nalls23andMe_2019/BST1/LD/BST1.1KGphase3_LD.RDS## Removing 1 temp files.## + FILTER:: Filtering by LD features.## ## ────────────────────────────────────────────────────────────────────────────────## ## ── Step 3 ▶▶▶ Filter SNPs 🚰 ───────────────────────────────────────────────────## ## ────────────────────────────────────────────────────────────────────────────────## FILTER:: Filtering by SNP features.## + FILTER:: Post-filtered data: 17 x 16## + Subsetting LD matrix and dat to common SNPs...## Removing unnamed rows/cols## Replacing NAs with 0## + LD_matrix = 17 SNPs.## + dat = 17 SNPs.## + 17 SNPs in common.## Converting obj to sparseMatrix.## ## ────────────────────────────────────────────────────────────────────────────────## ## ── Step 4 ▶▶▶ Fine-map 🔊 ──────────────────────────────────────────────────────## ## ────────────────────────────────────────────────────────────────────────────────## Gathering method sources.## Gathering method citations.## Preparing sample size column (N).## Using existing 'N' column.## Gathering method sources.## Gathering method citations.## ABF
## ✅ All required columns present.
## SUSIE
## ✅ All required columns present.
## ✅ All optional columns present.
## FINEMAP
## ✅ All required columns present.
## ✅ All optional columns present.## ++ Fine-mapping using 3 tool(s): ABF, SUSIE, FINEMAP##
## +++ Multi-finemap:: ABF +++## Using all default values for finemap_args$ABF## Preparing sample size column (N).## Using existing 'N' column.## Running ABF.## ++ Credible Set SNPs identified = 0## ++ Merging ABF results with multi-finemap data.##
## +++ Multi-finemap:: SUSIE +++## Using all default values for finemap_args$SUSIE## Loading required namespace: Rfast## Preparing sample size column (N).## Using existing 'N' column.## + SUSIE:: sample_size=1,474,097## + Subsetting LD matrix and dat to common SNPs...## Removing unnamed rows/cols## Replacing NAs with 0## + LD_matrix = 17 SNPs.## + dat = 17 SNPs.## + 17 SNPs in common.## Converting obj to sparseMatrix.## + SUSIE:: Using `susie_rss()` from susieR v0.12.27## + SUSIE:: Extracting Credible Sets.## ++ Credible Set SNPs identified = 0## ++ Merging SUSIE results with multi-finemap data.##
## +++ Multi-finemap:: FINEMAP +++## Using all default values for finemap_args$FINEMAP## Preparing sample size column (N).## Using existing 'N' column.## + Subsetting LD matrix and dat to common SNPs...## Removing unnamed rows/cols## Replacing NAs with 0## + LD_matrix = 17 SNPs.## + dat = 17 SNPs.## + 17 SNPs in common.## Converting obj to sparseMatrix.## Constructing master file.## Constructing data.z file.## Constructing data.ld file.## FINEMAP path: /github/home/.cache/R/echofinemap/FINEMAP/finemap_v1.4.1_x86_64/finemap_v1.4.1_x86_64## Inferred FINEMAP version: 1.4.1## Running FINEMAP.## cd .../BST1 &&
## .../finemap_v1.4.1_x86_64
##
## --sss
##
## --in-files .../master
##
## --log
##
## --n-causal-snps 5
##
## |--------------------------------------|
## | Welcome to FINEMAP v1.4.1 |
## | |
## | (c) 2015-2022 University of Helsinki |
## | |
## | Help : |
## | - ./finemap --help |
## | - www.finemap.me |
## | - www.christianbenner.com |
## | |
## | Contact : |
## | - finemap@christianbenner.com |
## | - matti.pirinen@helsinki.fi |
## |--------------------------------------|
##
## --------
## SETTINGS
## --------
## - dataset : all
## - corr-config : 0.95
## - n-causal-snps : 5
## - n-configs-top : 50000
## - n-conv-sss : 100
## - n-iter : 100000
## - n-threads : 1
## - prior-k0 : 0
## - prior-std : 0.05
## - prob-conv-sss-tol : 0.001
## - prob-cred-set : 0.95
##
## ------------
## FINE-MAPPING (1/1)
## ------------
## - GWAS summary stats : FINEMAP/data.z
## - SNP correlations : FINEMAP/data.ld
## - Causal SNP stats : FINEMAP/data.snp
## - Causal configurations : FINEMAP/data.config
## - Credible sets : FINEMAP/data.cred
## - Log file : FINEMAP/data.log_sss
##
- Reading summary statistics : +
- Reading summary statistics : -
- Reading summary statistics : +
- Reading summary stats : done!
- Reading SNP correlations : 0.000%
- Reading SNP correlations : done!
- Reading input : done!
##
## - Number of GWAS samples : 1474097
## - Number of SNPs : 17
## - Prior-Pr(# of causal SNPs is k) :
## (0 -> 0)
## 1 -> 0.59
## 2 -> 0.295
## 3 -> 0.0921
## 4 -> 0.0202
## 5 -> 0.00327
## 0 configurations evaluated (0.000/100%)
- 17 configurations evaluated (0.000/100%)
- 32 configurations evaluated (0.000/100%)
- 241 configurations evaluated (0.050/100%)
- 421 configurations evaluated (0.100/100%)
- 461 configurations evaluated (0.150/100%)
- 604 configurations evaluated (0.200/100%)
- 640 configurations evaluated (0.250/100%)
- 640 configurations evaluated (0.272/100%) : converged after 272 iterations
##
- Computing causal SNP statistics : 0.156%
- Computing causal SNP statistics : 7.969%
- Computing causal SNP statistics : 15.781%
- Computing causal SNP statistics : 23.594%
- Computing causal SNP statistics : 31.406%
- Computing causal SNP statistics : 39.219%
- Computing causal SNP statistics : 47.031%
- Computing causal SNP statistics : 54.844%
- Computing causal SNP statistics : 62.656%
- Computing causal SNP statistics : 70.469%
- Computing causal SNP statistics : 78.281%
- Computing causal SNP statistics : 86.094%
- Computing causal SNP statistics : 93.906%
- Computing causal SNP statistics : done!
## - Regional SNP heritability : 0.000117 (SD: 1.79e-05 ; 95% CI: [8.57e-05,0.000156])
## - Log10-BF of >= one causal SNP : 28.5
## - Post-expected # of causal SNPs : 5
## - Post-Pr(# of causal SNPs is k) :
## (0 -> 0)
## 1 -> 8.28e-06
## 2 -> 0.000269
## 3 -> 0.000144
## 4 -> 0.000238
## 5 -> 0.999
## - Computing credible sets :
- Writing causal configurations : 0.156%
- Writing causal configurations : 7.969%
- Writing causal configurations : 15.781%
- Writing causal configurations : 23.594%
- Writing causal configurations : 31.406%
- Writing causal configurations : 39.219%
- Writing causal configurations : 47.031%
- Writing causal configurations : 54.844%
- Writing causal configurations : 62.656%
- Writing causal configurations : 70.469%
- Writing causal configurations : 78.281%
- Writing causal configurations : 86.094%
- Writing causal configurations : 93.906%
- Writing causal SNP statistics : 5.882%
- Writing causal SNP statistics : 64.706%
- Writing output : done!
## - Run time : 0 hours, 0 minutes, 0 seconds## 1 data.cred* file(s) found in the same subfolder.## Selected file based on postPr_k: data.cred5## Importing conditional probabilities (.cred file).## Importing marginal probabilities (.snp file).## Importing configuration probabilities (.config file).## ++ Credible Set SNPs identified = 12## ++ Merging FINEMAP results with multi-finemap data.## Identifying Consensus SNPs...## + support_thresh = 2## + Calculating mean Posterior Probability (mean.PP)...## + 3 fine-mapping methods used.## + 12 Credible Set SNPs identified.## + 0 Consensus SNPs identified.## Saving merged finemapping results ==> /tmp/RtmpKaL0Dw/results/GWAS/Nalls23andMe_2019/BST1/Multi-finemap/1KGphase3_LD.Multi-finemap.tsv.gz## + Fine-mapping with 'ABF, SUSIE, FINEMAP' completed:## ## ────────────────────────────────────────────────────────────────────────────────## ## ── Step 5 ▶▶▶ Plot 📈 ──────────────────────────────────────────────────────────## ## ────────────────────────────────────────────────────────────────────────────────## +-------- Locus Plot: BST1 --------+## + support_thresh = 2## + Calculating mean Posterior Probability (mean.PP)...## + 3 fine-mapping methods used.## + 12 Credible Set SNPs identified.## + 0 Consensus SNPs identified.## + Filling NAs in CS cols with 0.## + Filling NAs in PP cols with 0.## LD_matrix detected. Coloring SNPs by LD with lead SNP.## ++ echoplot:: GWAS full window track## ++ echoplot:: GWAS track## ++ echoplot:: Merged fine-mapping track## Melting PP and CS from 4 fine-mapping methods.## + echoplot:: Constructing SNP labels.## Adding SNP group labels to locus plot.## ++ echoplot:: Adding Gene model track.## Converting dat to GRanges object.## Loading required namespace: EnsDb.Hsapiens.v75## max_transcripts= 1 .## 1 transcripts from 1 genes returned.## Fetching data...OK
## Parsing exons...OK
## Defining introns...OK
## Defining UTRs...OK
## Defining CDS...OK
## aggregating...
## Done
## Constructing graphics...
## + Adding vertical lines to highlight SNP groups.
## +>+>+>+>+ zoom = 1x +<+<+<+<+
## + echoplot:: Get window suffix...
## + echoplot:: Removing GWAS full window track @ zoom=1x
## + Removing subplot margins...
## + Reordering tracks...
## + Ensuring last track shows genomic units.
## + Aligning xlimits for each subplot...
## + Checking track heights...
## + echoplot:: Saving plot ==> /tmp/RtmpKaL0Dw/results/GWAS/Nalls23andMe_2019/BST1/multiview.BST1.1KGphase3.1x.png
## Found more than one class "simpleUnit" in cache; using the first, from namespace 'hexbin'
## Also defined by 'ggbio'
## Recording all `finemap_locus` arguments.
## Formatting locus results.## ┌─────────────────────────────────────────┐
## │ │
## │ )))> 🦇 MEX3C [locus 2 / 2] 🦇 <((( │
## │ │
## └─────────────────────────────────────────┘##
## ────────────────────────────────────────────────────────────────────────────────
##
## ── Step 1 ▶▶▶ Query 🔎 ─────────────────────────────────────────────────────────
##
## ────────────────────────────────────────────────────────────────────────────────
## + Query Method: tabix
## Constructing GRanges query using min/max ranges within a single chromosome.
## query_dat is already a GRanges object. Returning directly.
## ========= echotabix::convert =========
## Converting full summary stats file to tabix format for fast querying.
## Inferred format: 'table'
## Explicit format: 'table'
## Inferring comment_char from tabular header: 'SNP'
## Determining chrom type from file header.
## Chromosome format: 1
## Detecting column delimiter.
## Identified column separator: \t
## Sorting rows by coordinates via bash.
## Searching for header row with grep.## ( grep ^'SNP' .../nalls2019.fullSS_subset.tsv; grep
## -v ^'SNP' .../nalls2019.fullSS_subset.tsv | sort
## -k2,2n
## -k3,3n ) > .../file134ae6ba11efe_sorted.tsv## Constructing outputs
## Using existing bgzipped file: /tmp/RtmpKaL0Dw/nalls2019.fullSS_subset.tsv.bgz
## Set force_new=TRUE to override this.
## Tabix-indexing file using: Rsamtools
## Data successfully converted to bgzip-compressed, tabix-indexed format.
## ========= echotabix::query =========
## query_dat is already a GRanges object. Returning directly.
## Inferred format: 'table'
## Querying tabular tabix file using: Rsamtools.
## Checking query chromosome style is correct.
## Chromosome format: 1
## Retrieving data.
## Converting query results to data.table.
## Processing query: 18:48682589-48684589
## Adding 'query' column to results.
## Retrieved data with 7 rows
## Saving query ==> /tmp/RtmpKaL0Dw/results/GWAS/Nalls23andMe_2019/MEX3C/MEX3C_Nalls23andMe_2019_subset.tsv.gz
## + Query: 7 SNPs x 11 columns.
## Standardizing summary statistics subset.
## Standardizing main column names.
## ++ Preparing A1,A1 cols
## ++ Preparing MAF,Freq cols.
## ++ Could not infer MAF from Freq.
## ++ Preparing N_cases,N_controls cols.
## ++ Preparing proportion_cases col.
## ++ Calculating proportion_cases from N_cases and N_controls.
## Preparing sample size column (N).
## Computing effective sample size using the LDSC method:
## Neff = (N_CAS+N_CON) * (N_CAS/(N_CAS+N_CON)) / mean((N_CAS/(N_CAS+N_CON))[(N_CAS+N_CON)==max(N_CAS+N_CON)]))
## + Mapping colnames from MungeSumstats ==> echolocatoR
## + Imputing t-statistic from Effect and StdErr.
## + leadSNP missing. Assigning new one by min p-value.
## ++ Ensuring Effect,StdErr,P are numeric.
## ++ Ensuring 1 SNP per row and per genomic coordinate.
## ++ Removing extra whitespace
## + Standardized query: 7 SNPs x 15 columns.
## ++ Saving standardized query ==> /tmp/RtmpKaL0Dw/results/GWAS/Nalls23andMe_2019/MEX3C/MEX3C_Nalls23andMe_2019_subset.tsv.gz
##
## ────────────────────────────────────────────────────────────────────────────────
##
## ── Step 2 ▶▶▶ Extract Linkage Disequilibrium 🔗 ────────────────────────────────
##
## ────────────────────────────────────────────────────────────────────────────────
## LD_reference identified as: 1kg.
## Using 1000Genomes as LD reference panel.
## Constructing GRanges query using min/max ranges across one or more chromosomes.
## + as_blocks=TRUE: Will query a single range per chromosome that covers all regions requested (plus anything in between).
## LD Reference Panel = 1KGphase3
## Querying 1KG remote server.
## Selecting 503 EUR individuals from 1kgphase3.
## ========= echotabix::query =========
## query_dat is already a GRanges object. Returning directly.
## Explicit format: 'vcf'
## Querying VCF tabix file.
## Querying VCF file using: VariantAnnotation
## Checking query chromosome style is correct.
## Chromosome format: 1
## Filtering query to 503 samples and returning ScanVcfParam object.
## Retrieving data.
## Time difference of 4.2 secs
## Removing 7 / 38 non-overlapping SNPs.
## Saving VCF subset ==> /tmp/RtmpKaL0Dw/VCF/RtmpKaL0Dw.chr18-48682662-48684103.ALL.chr18.phase3_shapeit2_mvncall_integrated_v5a.20130502.genotypes.vcf.bgz## Warning in .Call(.make_vcf_geno, filename, fixed, names(geno), as.list(geno), :
## converting NULL pointer to R NULL## Time difference of 0.3 secs
## Retrieved data with 7 rows across 503 samples.
## echoLD::snpStats:: Filling `MAF` column with MAF from LD panel.
## echoLD:snpStats:: Computing pairwise LD between 7 SNPs across 503 individuals (stats = R).
## Time difference of 0 secs
## 7 x 7 LD_matrix (sparse)
## Converting obj to sparseMatrix.
## Saving sparse LD matrix ==> /tmp/RtmpKaL0Dw/results/GWAS/Nalls23andMe_2019/MEX3C/LD/MEX3C.1KGphase3_LD.RDS
## Removing 1 temp files.
## + FILTER:: Filtering by LD features.
##
## ────────────────────────────────────────────────────────────────────────────────
##
## ── Step 3 ▶▶▶ Filter SNPs 🚰 ───────────────────────────────────────────────────
##
## ────────────────────────────────────────────────────────────────────────────────
## FILTER:: Filtering by SNP features.
## + FILTER:: Post-filtered data: 5 x 16
## + Subsetting LD matrix and dat to common SNPs...
## Removing unnamed rows/cols
## Replacing NAs with 0
## + LD_matrix = 7 SNPs.
## + dat = 5 SNPs.
## + 5 SNPs in common.
## Converting obj to sparseMatrix.
##
## ────────────────────────────────────────────────────────────────────────────────
##
## ── Step 4 ▶▶▶ Fine-map 🔊 ──────────────────────────────────────────────────────
##
## ────────────────────────────────────────────────────────────────────────────────
## Gathering method sources.
## Gathering method citations.
## Preparing sample size column (N).
## Using existing 'N' column.
## Gathering method sources.
## Gathering method citations.## ABF
## ✅ All required columns present.
## SUSIE
## ✅ All required columns present.
## ✅ All optional columns present.
## FINEMAP
## ✅ All required columns present.
## ✅ All optional columns present.## ++ Fine-mapping using 3 tool(s): ABF, SUSIE, FINEMAP
##
## +++ Multi-finemap:: ABF +++
## Using all default values for finemap_args$ABF
## Preparing sample size column (N).
## Using existing 'N' column.
## Running ABF.
## ++ Credible Set SNPs identified = 0
## ++ Merging ABF results with multi-finemap data.
##
## +++ Multi-finemap:: SUSIE +++
## Using all default values for finemap_args$SUSIE
## Preparing sample size column (N).
## Using existing 'N' column.
## + SUSIE:: sample_size=1,460,059
## + Subsetting LD matrix and dat to common SNPs...
## Removing unnamed rows/cols
## Replacing NAs with 0
## + LD_matrix = 5 SNPs.
## + dat = 5 SNPs.
## + 5 SNPs in common.
## Converting obj to sparseMatrix.
## + SUSIE:: Using `susie_rss()` from susieR v0.12.27
## + SUSIE:: Extracting Credible Sets.
## ++ Credible Set SNPs identified = 0
## ++ Merging SUSIE results with multi-finemap data.
##
## +++ Multi-finemap:: FINEMAP +++
## Using all default values for finemap_args$FINEMAP
## Preparing sample size column (N).
## Using existing 'N' column.
## + Subsetting LD matrix and dat to common SNPs...
## Removing unnamed rows/cols
## Replacing NAs with 0
## + LD_matrix = 5 SNPs.
## + dat = 5 SNPs.
## + 5 SNPs in common.
## Converting obj to sparseMatrix.
## Constructing master file.
## Constructing data.z file.
## Constructing data.ld file.
## FINEMAP path: /github/home/.cache/R/echofinemap/FINEMAP/finemap_v1.4.1_x86_64/finemap_v1.4.1_x86_64
## Inferred FINEMAP version: 1.4.1
## Running FINEMAP.## cd .../MEX3C &&
## .../finemap_v1.4.1_x86_64
##
## --sss
##
## --in-files .../master
##
## --log
##
## --n-causal-snps 5
##
## |--------------------------------------|
## | Welcome to FINEMAP v1.4.1 |
## | |
## | (c) 2015-2022 University of Helsinki |
## | |
## | Help : |
## | - ./finemap --help |
## | - www.finemap.me |
## | - www.christianbenner.com |
## | |
## | Contact : |
## | - finemap@christianbenner.com |
## | - matti.pirinen@helsinki.fi |
## |--------------------------------------|
##
## --------
## SETTINGS
## --------
## - dataset : all
## - corr-config : 0.95
## - n-causal-snps : 5
## - n-configs-top : 50000
## - n-conv-sss : 100
## - n-iter : 100000
## - n-threads : 1
## - prior-k0 : 0
## - prior-std : 0.05
## - prob-conv-sss-tol : 0.001
## - prob-cred-set : 0.95
##
## ------------
## FINE-MAPPING (1/1)
## ------------
## - GWAS summary stats : FINEMAP/data.z
## - SNP correlations : FINEMAP/data.ld
## - Causal SNP stats : FINEMAP/data.snp
## - Causal configurations : FINEMAP/data.config
## - Credible sets : FINEMAP/data.cred
## - Log file : FINEMAP/data.log_sss
##
- Reading summary statistics : +
- Reading summary stats : done!
- Reading SNP correlations : 0.000%
- Reading SNP correlations : done!
- Reading input : done!
##
## - Number of GWAS samples : 1460059
## - Number of SNPs : 5
## - Prior-Pr(# of causal SNPs is k) :
## (0 -> 0)
## 1 -> 0.609
## 2 -> 0.305
## 3 -> 0.0762
## 4 -> 0.00952
## 5 -> 0.000476
## 0 configurations evaluated (0.000/100%)
- 5 configurations evaluated (0.000/100%)
- 8 configurations evaluated (0.000/100%)
- 12 configurations evaluated (0.050/100%)
- 12 configurations evaluated (0.100/100%)
- 12 configurations evaluated (0.104/100%) : converged after 104 iterations
##
- Computing causal SNP statistics : 8.333%
- Computing causal SNP statistics : done!
## - Regional SNP heritability : 1.93e-05 (SD: 7.35e-06 ; 95% CI: [7.27e-06,3.53e-05])
## - Log10-BF of >= one causal SNP : 3.56
## - Post-expected # of causal SNPs : 1.09
## - Post-Pr(# of causal SNPs is k) :
## (0 -> 0)
## 1 -> 0.915
## 2 -> 0.0853
## 3 -> 0.000204
## 4 -> 0
## 5 -> 0
## - Computing credible sets :
- Writing causal configurations : 8.333%
- Writing causal SNP statistics : 20.000%
- Writing output : done!
## - Run time : 0 hours, 0 minutes, 0 seconds## 2 data.cred* file(s) found in the same subfolder.
## Selected file based on postPr_k: data.cred1
## Importing conditional probabilities (.cred file).
## No configurations were causal at PP>=0.95.
## Importing marginal probabilities (.snp file).
## Importing configuration probabilities (.config file).
## FINEMAP was unable to identify any credible sets at PP>=0.95.
## ++ Credible Set SNPs identified = 0
## ++ Merging FINEMAP results with multi-finemap data.
## Identifying Consensus SNPs...
## + support_thresh = 2
## + Calculating mean Posterior Probability (mean.PP)...
## + 3 fine-mapping methods used.
## + 0 Credible Set SNPs identified.
## + 0 Consensus SNPs identified.
## Saving merged finemapping results ==> /tmp/RtmpKaL0Dw/results/GWAS/Nalls23andMe_2019/MEX3C/Multi-finemap/1KGphase3_LD.Multi-finemap.tsv.gz
## + Fine-mapping with 'ABF, SUSIE, FINEMAP' completed:
##
## ────────────────────────────────────────────────────────────────────────────────
##
## ── Step 5 ▶▶▶ Plot 📈 ──────────────────────────────────────────────────────────
##
## ────────────────────────────────────────────────────────────────────────────────
## +-------- Locus Plot: MEX3C --------+
## + support_thresh = 2
## + Calculating mean Posterior Probability (mean.PP)...
## + 3 fine-mapping methods used.
## + 0 Credible Set SNPs identified.
## + 0 Consensus SNPs identified.
## + Filling NAs in CS cols with 0.
## + Filling NAs in PP cols with 0.
## LD_matrix detected. Coloring SNPs by LD with lead SNP.
## ++ echoplot:: GWAS full window track
## ++ echoplot:: GWAS track
## ++ echoplot:: Merged fine-mapping track
## Melting PP and CS from 4 fine-mapping methods.
## + echoplot:: Constructing SNP labels.
## Adding SNP group labels to locus plot.
## ++ echoplot:: Adding Gene model track.
## Converting dat to GRanges object.
## max_transcripts= 1 .
## 0 transcripts from 0 genes returned.
## Returning NULL for gene track.
## + Adding vertical lines to highlight SNP groups.
## +>+>+>+>+ zoom = 1x +<+<+<+<+
## + echoplot:: Get window suffix...
## + echoplot:: Removing GWAS full window track @ zoom=1x
## + Removing subplot margins...
## + Reordering tracks...
## + Ensuring last track shows genomic units.
## + Aligning xlimits for each subplot...
## + Checking track heights...
## + echoplot:: Saving plot ==> /tmp/RtmpKaL0Dw/results/GWAS/Nalls23andMe_2019/MEX3C/multiview.MEX3C.1KGphase3.1x.png## Warning: Removed 1 rows containing missing values (`geom_text()`).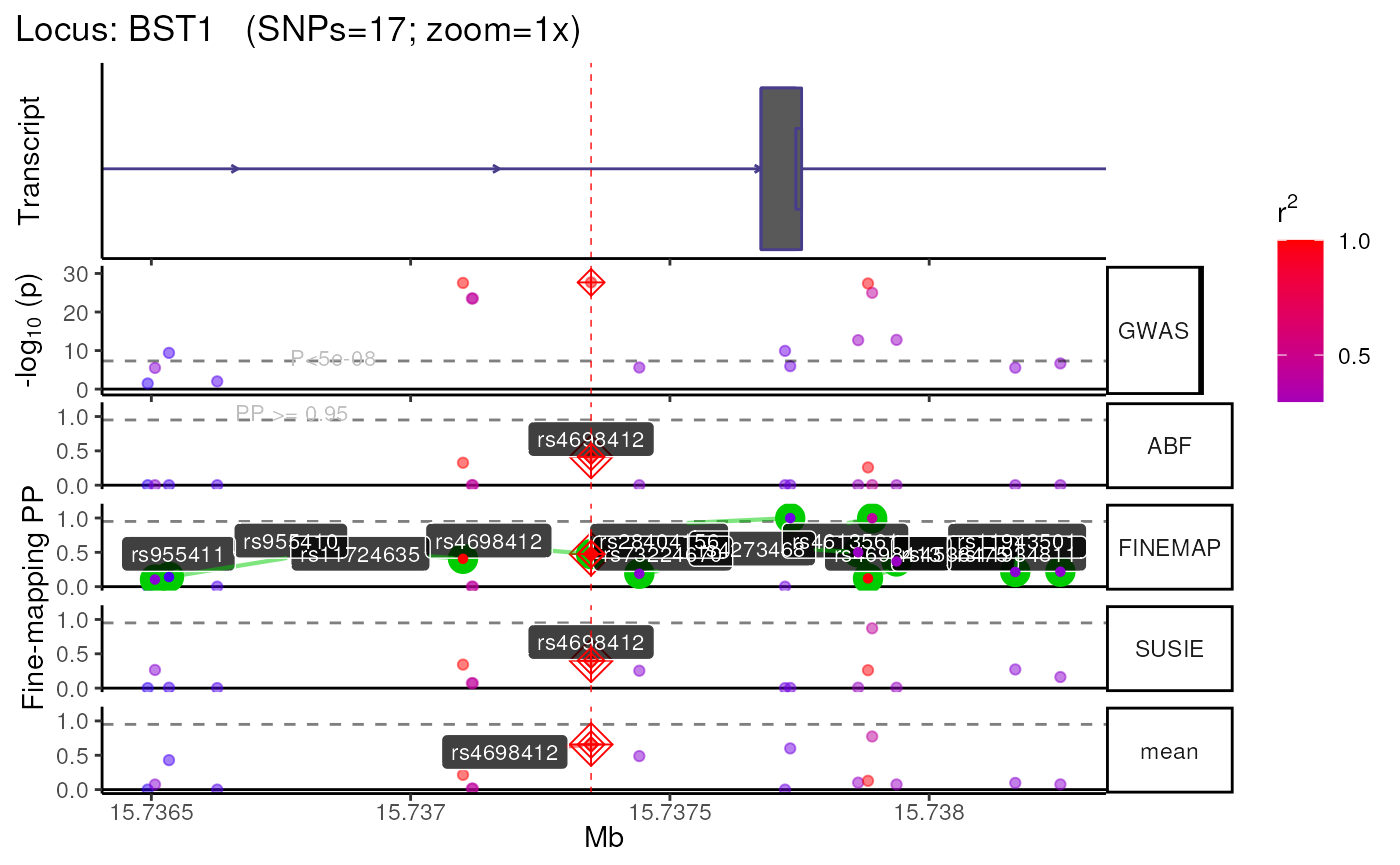
## Warning: Removed 1 rows containing missing values (`geom_text()`).## Recording all `finemap_locus` arguments.
## Formatting locus results.
##
## ────────────────────────────────────────────────────────────────────────────────
##
## ── Step 6 ▶▶▶ Postprocess data 🎁 ──────────────────────────────────────────────
##
## ────────────────────────────────────────────────────────────────────────────────
## Returning results as nested list.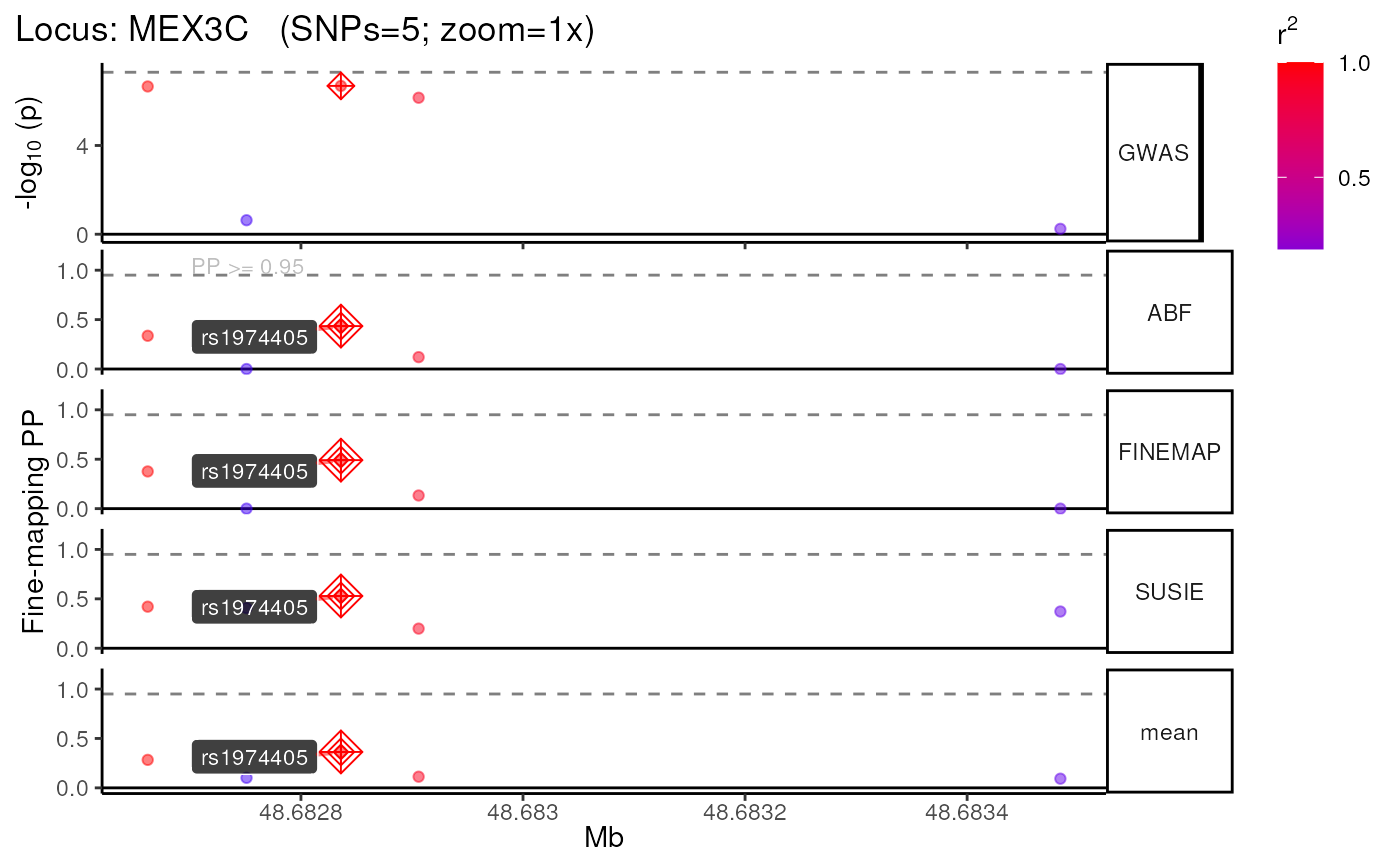
## All loci done in: 0.48 min
## Session info
utils::sessionInfo()## R Under development (unstable) (2023-01-11 r83598)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 22.04.1 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## time zone: UTC
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] snpStats_1.49.0 Matrix_1.5-3 survival_3.5-0 echolocatoR_2.0.3
## [5] BiocStyle_2.27.0
##
## loaded via a namespace (and not attached):
## [1] ProtGenerics_1.31.0 fs_1.5.2
## [3] matrixStats_0.63.0 bitops_1.0-7
## [5] EnsDb.Hsapiens.v75_2.99.0 httr_1.4.4
## [7] RColorBrewer_1.1-3 Rgraphviz_2.43.0
## [9] tools_4.3.0 backports_1.4.1
## [11] utf8_1.2.2 R6_2.5.1
## [13] DT_0.27 lazyeval_0.2.2
## [15] withr_2.5.0 prettyunits_1.1.1
## [17] GGally_2.1.2 gridExtra_2.3
## [19] cli_3.6.0 Biobase_2.59.0
## [21] textshaping_0.3.6 labeling_0.4.2
## [23] ggbio_1.47.0 sass_0.4.4
## [25] mvtnorm_1.1-3 readr_2.1.3
## [27] proxy_0.4-27 pkgdown_2.0.7
## [29] mixsqp_0.3-48 Rsamtools_2.15.1
## [31] systemfonts_1.0.4 foreign_0.8-84
## [33] R.utils_2.12.2 dichromat_2.0-0.1
## [35] maps_3.4.1 BSgenome_1.67.3
## [37] readxl_1.4.1 susieR_0.12.27
## [39] pals_1.7 rstudioapi_0.14
## [41] RSQLite_2.2.20 httpcode_0.3.0
## [43] generics_0.1.3 BiocIO_1.9.1
## [45] echoconda_0.99.9 dplyr_1.0.10
## [47] zip_2.2.2 interp_1.1-3
## [49] fansi_1.0.3 DescTools_0.99.47
## [51] S4Vectors_0.37.3 catalogueR_1.0.1
## [53] R.methodsS3_1.8.2 lifecycle_1.0.3
## [55] yaml_2.3.6 SummarizedExperiment_1.29.1
## [57] BiocFileCache_2.7.1 echoplot_0.99.6
## [59] grid_4.3.0 blob_1.2.3
## [61] crayon_1.5.2 dir.expiry_1.7.0
## [63] lattice_0.20-45 GenomicFeatures_1.51.2
## [65] mapproj_1.2.11 KEGGREST_1.39.0
## [67] MungeSumstats_1.7.15 pillar_1.8.1
## [69] knitr_1.41 GenomicRanges_1.51.4
## [71] rjson_0.2.21 osfr_0.2.9
## [73] boot_1.3-28.1 gld_2.6.6
## [75] codetools_0.2-18 glue_1.6.2
## [77] data.table_1.14.6 coloc_5.1.0.1
## [79] vctrs_0.5.1 png_0.1-8
## [81] XGR_1.1.8 cellranger_1.1.0
## [83] gtable_0.3.1 assertthat_0.2.1
## [85] cachem_1.0.6 dnet_1.1.7
## [87] xfun_0.36 openxlsx_4.2.5.1
## [89] Rfast_2.0.6 gargle_1.2.1
## [91] ellipsis_0.3.2 nlme_3.1-161
## [93] bit64_4.0.5 progress_1.2.2
## [95] filelock_1.0.2 googleAuthR_2.0.0
## [97] GenomeInfoDb_1.35.12 rprojroot_2.0.3
## [99] bslib_0.4.2 irlba_2.3.5.1
## [101] rpart_4.1.19 colorspace_2.0-3
## [103] BiocGenerics_0.45.0 DBI_1.1.3
## [105] Hmisc_4.7-2 nnet_7.3-18
## [107] Exact_3.2 tidyselect_1.2.0
## [109] bit_4.0.5 compiler_4.3.0
## [111] curl_5.0.0 graph_1.77.1
## [113] htmlTable_2.4.1 expm_0.999-7
## [115] basilisk.utils_1.11.1 xml2_1.3.3
## [117] desc_1.4.2 DelayedArray_0.25.0
## [119] bookdown_0.32 rtracklayer_1.59.1
## [121] checkmate_2.1.0 scales_1.2.1
## [123] hexbin_1.28.2 echoLD_0.99.9
## [125] RBGL_1.75.0 RCircos_1.2.2
## [127] rappdirs_0.3.3 stringr_1.5.0
## [129] supraHex_1.37.0 digest_0.6.31
## [131] piggyback_0.1.4 rmarkdown_2.20
## [133] basilisk_1.11.2 XVector_0.39.0
## [135] htmltools_0.5.4 pkgconfig_2.0.3
## [137] jpeg_0.1-10 base64enc_0.1-3
## [139] MatrixGenerics_1.11.0 echodata_0.99.16
## [141] highr_0.10 ensembldb_2.23.1
## [143] dbplyr_2.3.0 fastmap_1.1.0
## [145] rlang_1.0.6 htmlwidgets_1.6.1
## [147] farver_2.1.1 echofinemap_0.99.5
## [149] jquerylib_0.1.4 jsonlite_1.8.4
## [151] BiocParallel_1.33.9 R.oo_1.25.0
## [153] VariantAnnotation_1.45.0 RCurl_1.98-1.9
## [155] magrittr_2.0.3 Formula_1.2-4
## [157] GenomeInfoDbData_1.2.9 ggnetwork_0.5.10
## [159] patchwork_1.1.2 munsell_0.5.0
## [161] Rcpp_1.0.9 ggnewscale_0.4.8
## [163] ape_5.6-2 viridis_0.6.2
## [165] reticulate_1.27 RcppZiggurat_0.1.6
## [167] stringi_1.7.12 rootSolve_1.8.2.3
## [169] zlibbioc_1.45.0 MASS_7.3-58.1
## [171] plyr_1.8.8 parallel_4.3.0
## [173] ggrepel_0.9.2 lmom_2.9
## [175] deldir_1.0-6 echoannot_0.99.10
## [177] Biostrings_2.67.0 splines_4.3.0
## [179] hms_1.1.2 igraph_1.3.5
## [181] reshape2_1.4.4 biomaRt_2.55.0
## [183] stats4_4.3.0 crul_1.3
## [185] XML_3.99-0.13 evaluate_0.20
## [187] latticeExtra_0.6-30 biovizBase_1.47.0
## [189] BiocManager_1.30.19 tzdb_0.3.0
## [191] tidyr_1.2.1 purrr_1.0.1
## [193] reshape_0.8.9 ggplot2_3.4.0
## [195] echotabix_0.99.9 restfulr_0.0.15
## [197] AnnotationFilter_1.23.0 e1071_1.7-12
## [199] downloadR_0.99.6 viridisLite_0.4.1
## [201] class_7.3-20.1 ragg_1.2.5
## [203] OrganismDbi_1.41.0 tibble_3.1.8
## [205] memoise_2.0.1 AnnotationDbi_1.61.0
## [207] GenomicAlignments_1.35.0 IRanges_2.33.0
## [209] cluster_2.1.4