# knitr::opts_chunk$set(error = T) # Knit even with errors
# source("echolocatoR/R/MAIN.R")
library(dplyr)
library(stringr); library(bsselectR) # devtools::install_github("walkerke/bsselectR")Summarize Fine-mapping Results
# .tabset-dropdown
loci <- list.dirs("./Data/GWAS/Nalls23andMe_2019", recursive = F) %>% basename()
loci <- loci[loci!="_genome_wide"]
plot.paths <- file.path("./Data/GWAS/Nalls23andMe_2019",
loci, "Multi-finemap",
paste0(loci,"_ggbio.png"))
# Add link to plot
# baseURL <- "https://github.com/RajLabMSSM/Fine_Mapping/blob/master/Data/GWAS/Nalls23andMe_2019"
# links <- data.frame(Link=file.path(baseURL, top_loci_sort$Locus,"Multi-finemap",
# paste0(top_loci_sort$Locus,"_ggbio.png")))
# top_loci_sort <- cbind(links, top_loci_sort)
# file.path("https://rajlabmssm.github.io/Fine_Mapping/Data/Nalls23andMe_2019",
# genes,"Multi-finemap", paste0(genes,"_ggbio.png"))
# Dropdown
# genes <- basename(dirname(dirname(plot.paths)))
# plot.paths <- file.path("https://rajlabmssm.github.io/Fine_Mapping/Data/GWAS/Nalls23andMe_2019",genes,"Multi-finemap",paste0(genes,"_ggbio.png"))
# names(plot.paths) <- genes
#
#
# bsselect(vector = plot.paths,
# type = "img",
# selected = "LRRK2",
# frame_height = "1000",
# live_search = T,
# show_tick = T)
for (p in plot.paths){
if(file.exists(p)){
try({
gene <- basename(dirname(dirname(p)))
cat(' \n###', gene, ' \n')
cat(paste0(''))
cat(' \n')
})
}
}##
## ### ASXL3
## 
##
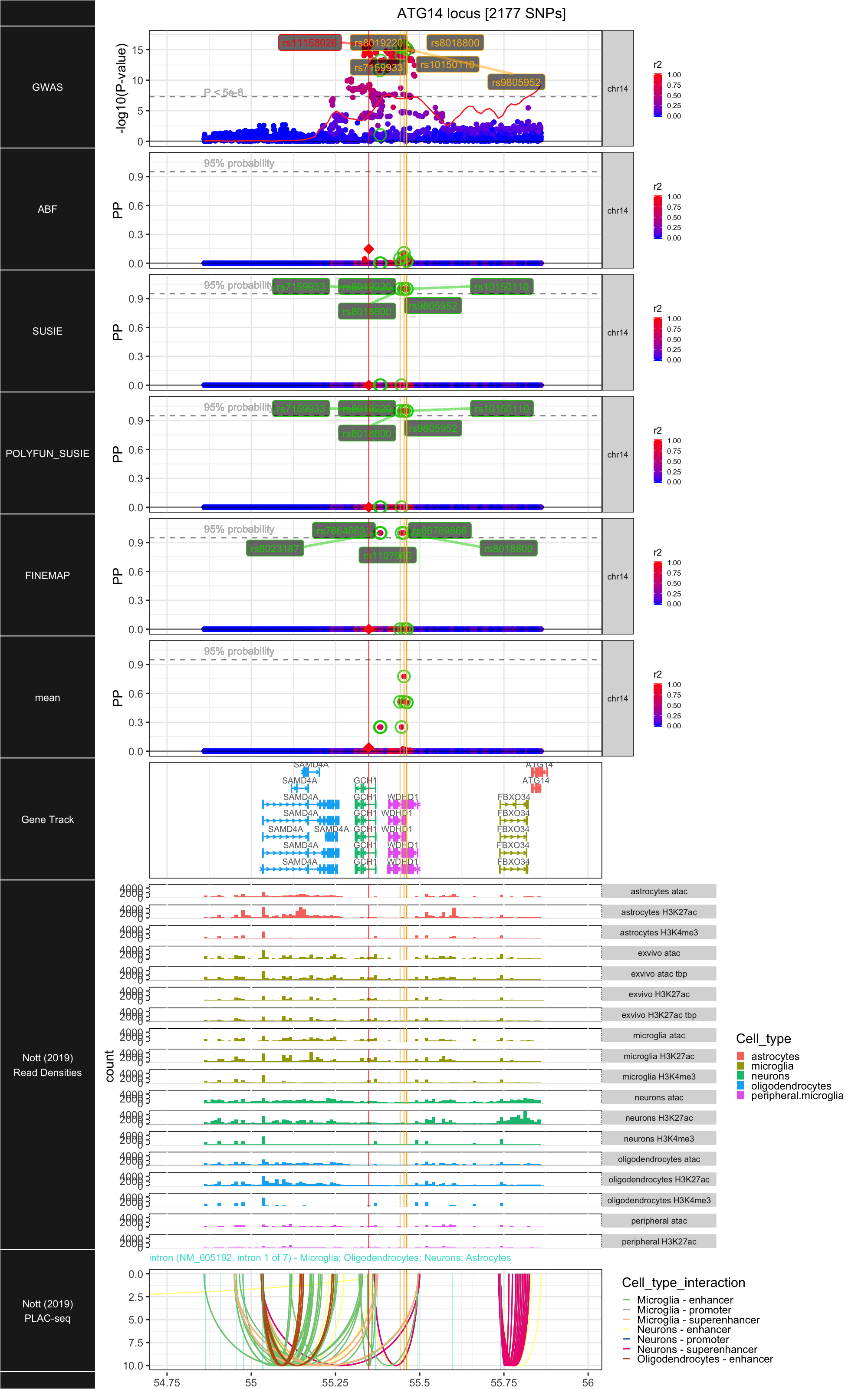
## ### ATG14
## 
##
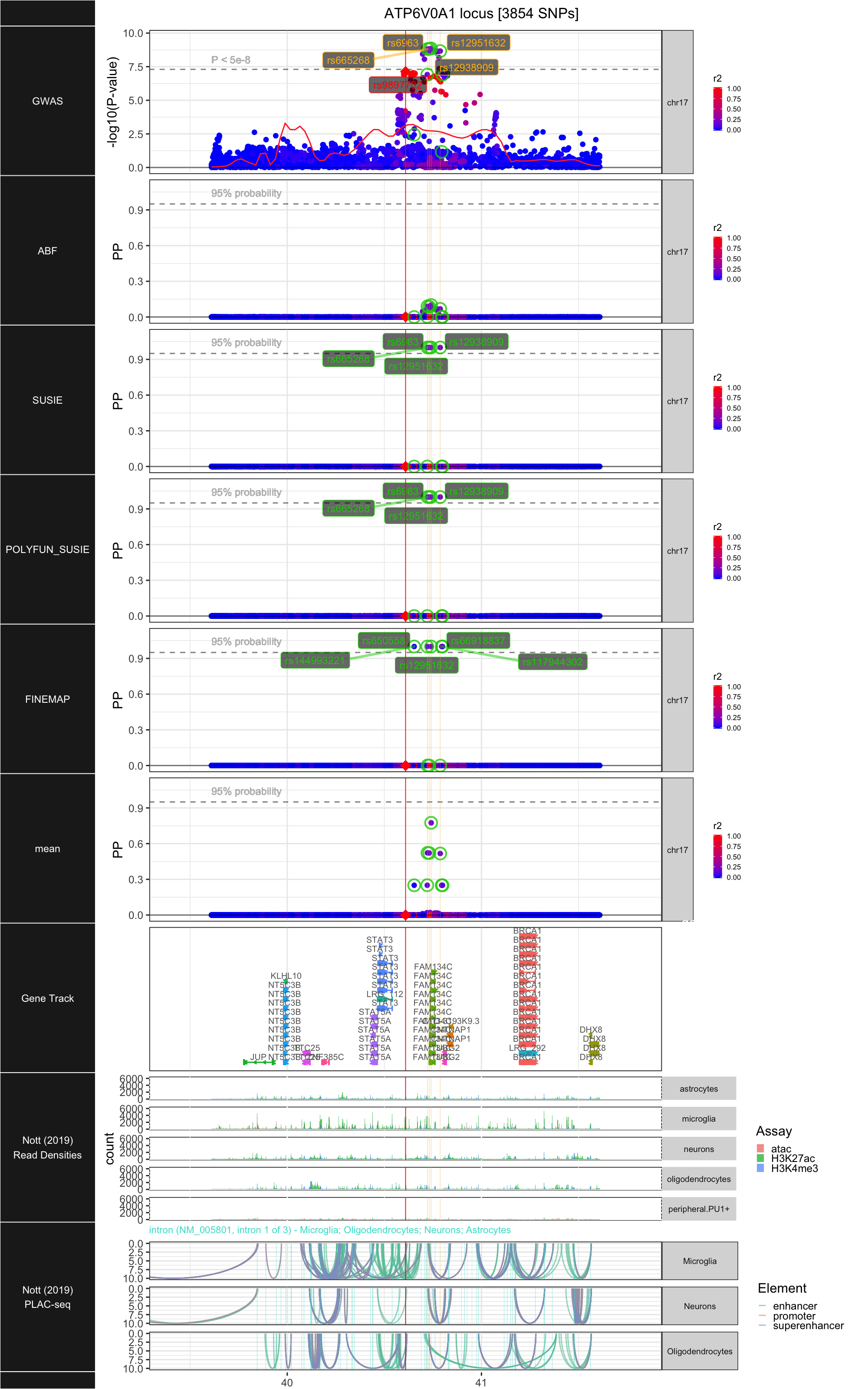
## ### ATP6V0A1
## 
##
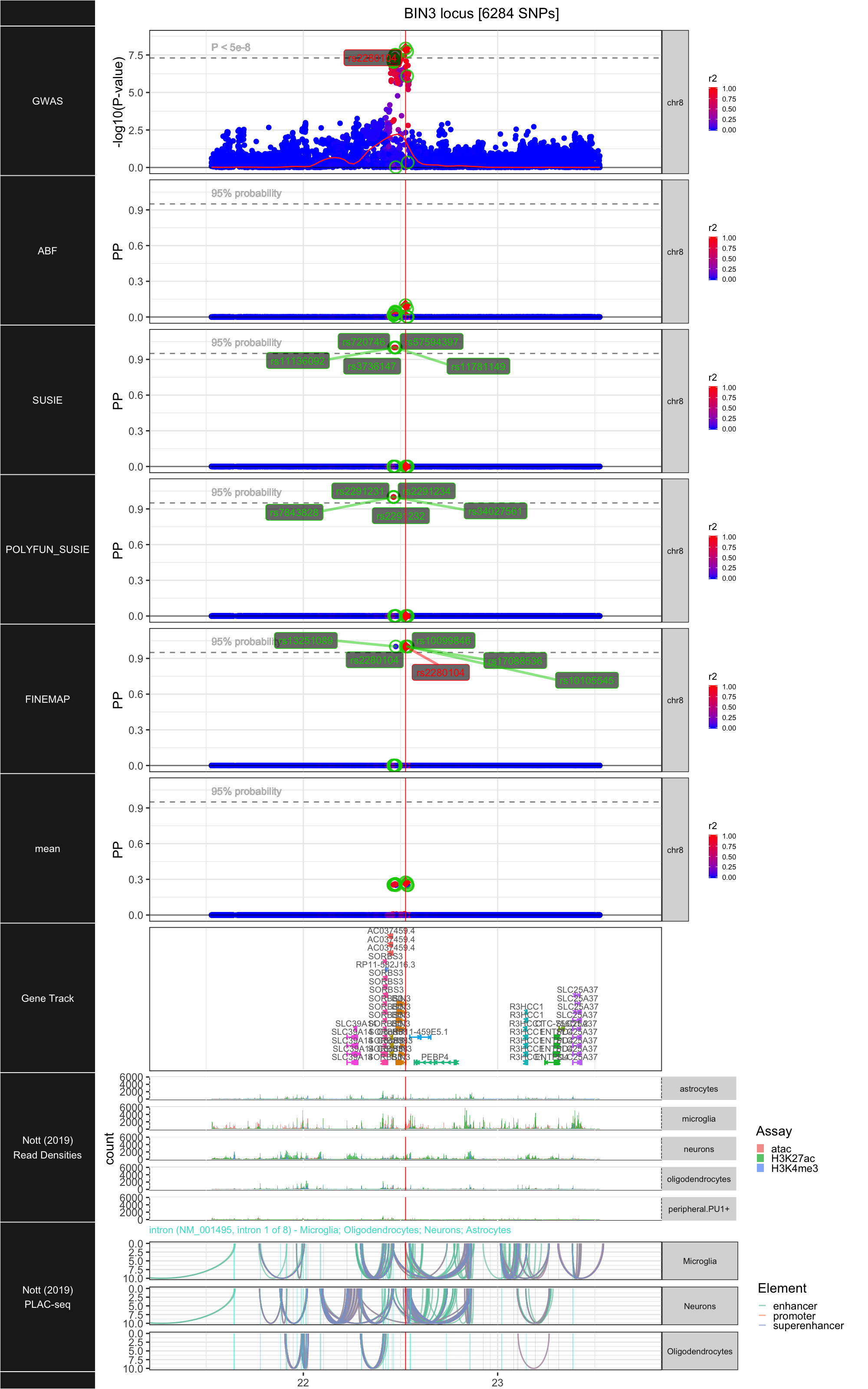
## ### BIN3
## 
##
## ### BRIP1
## 
##
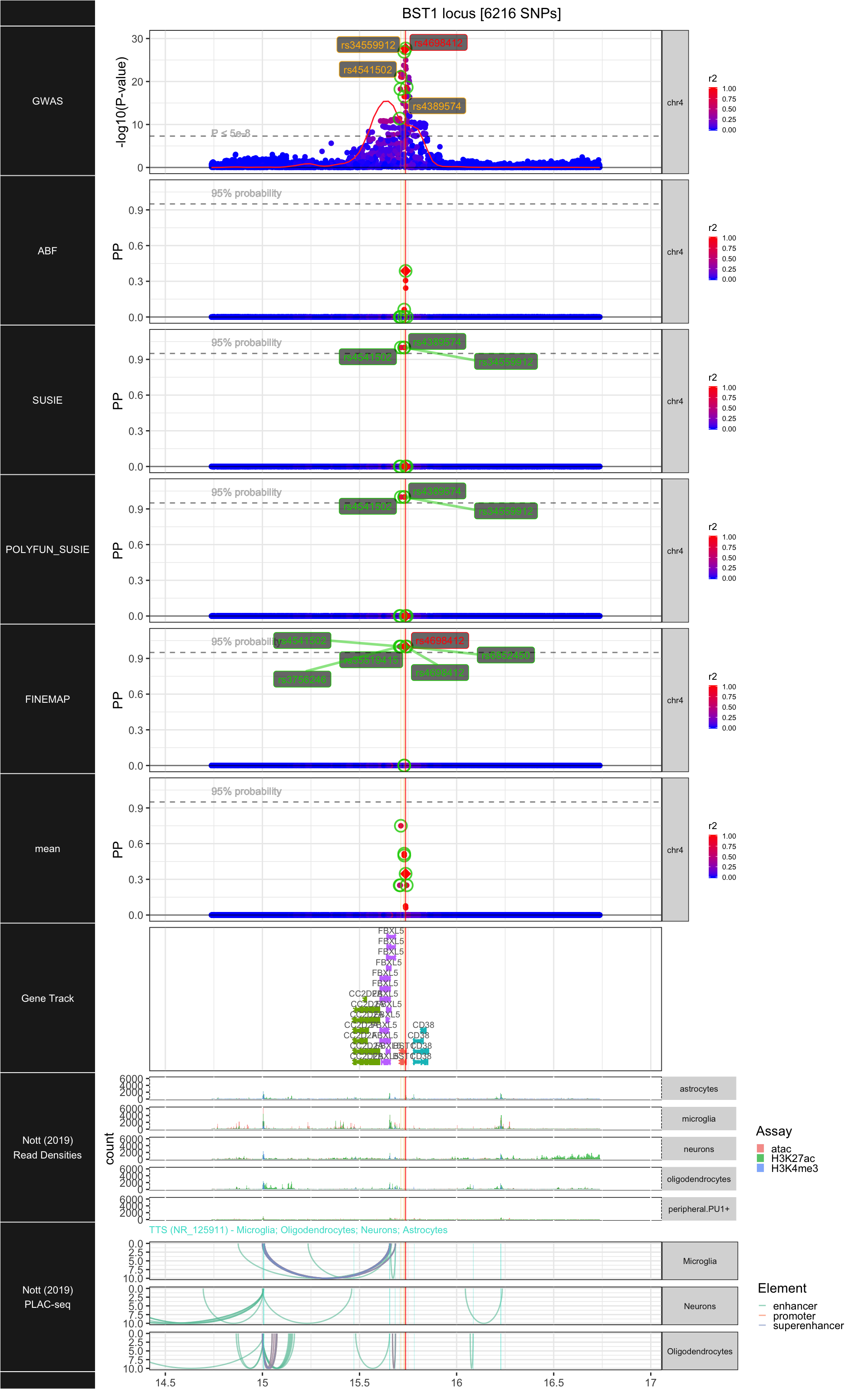
## ### BST1
## 
##
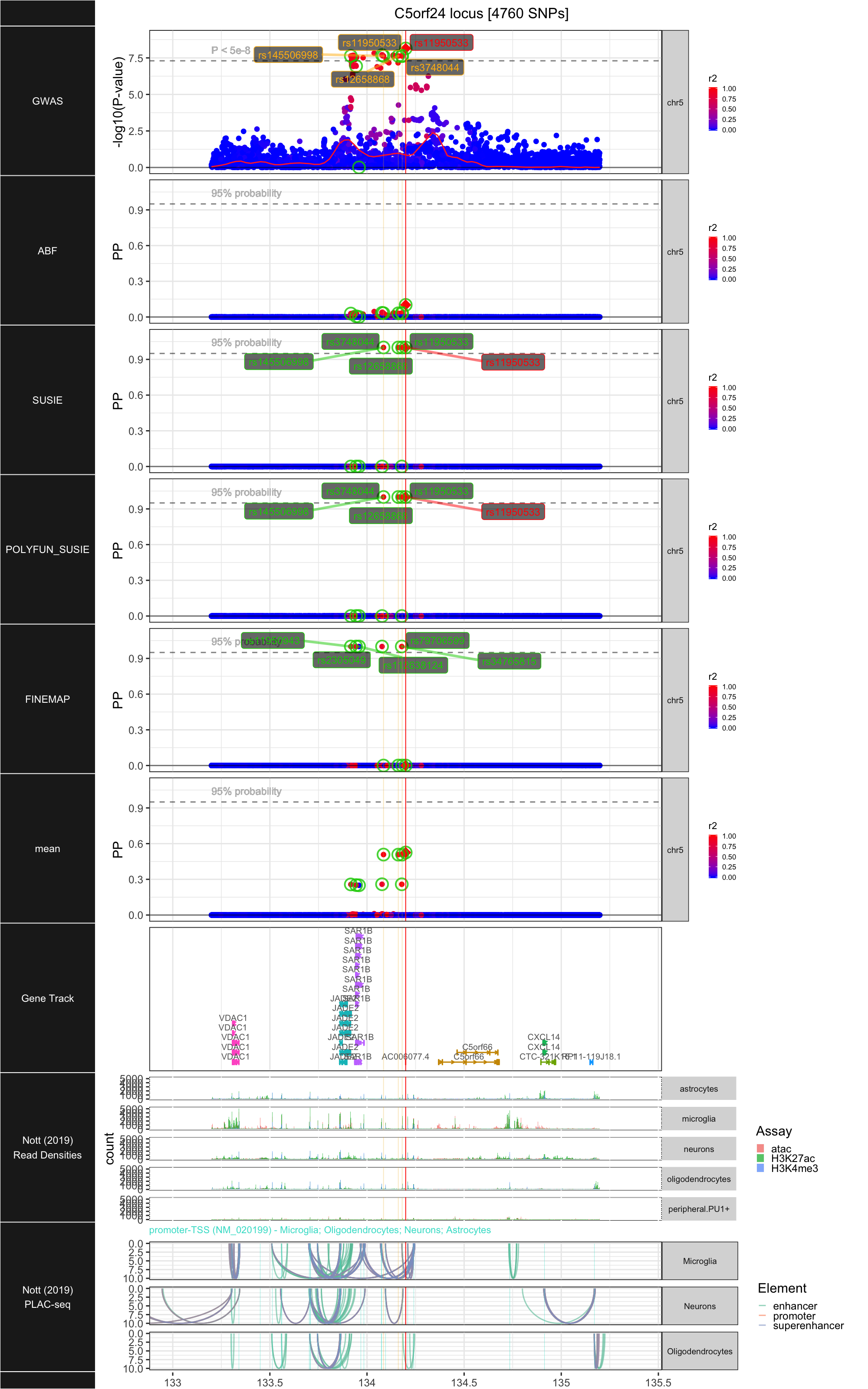
## ### C5orf24
## 
##
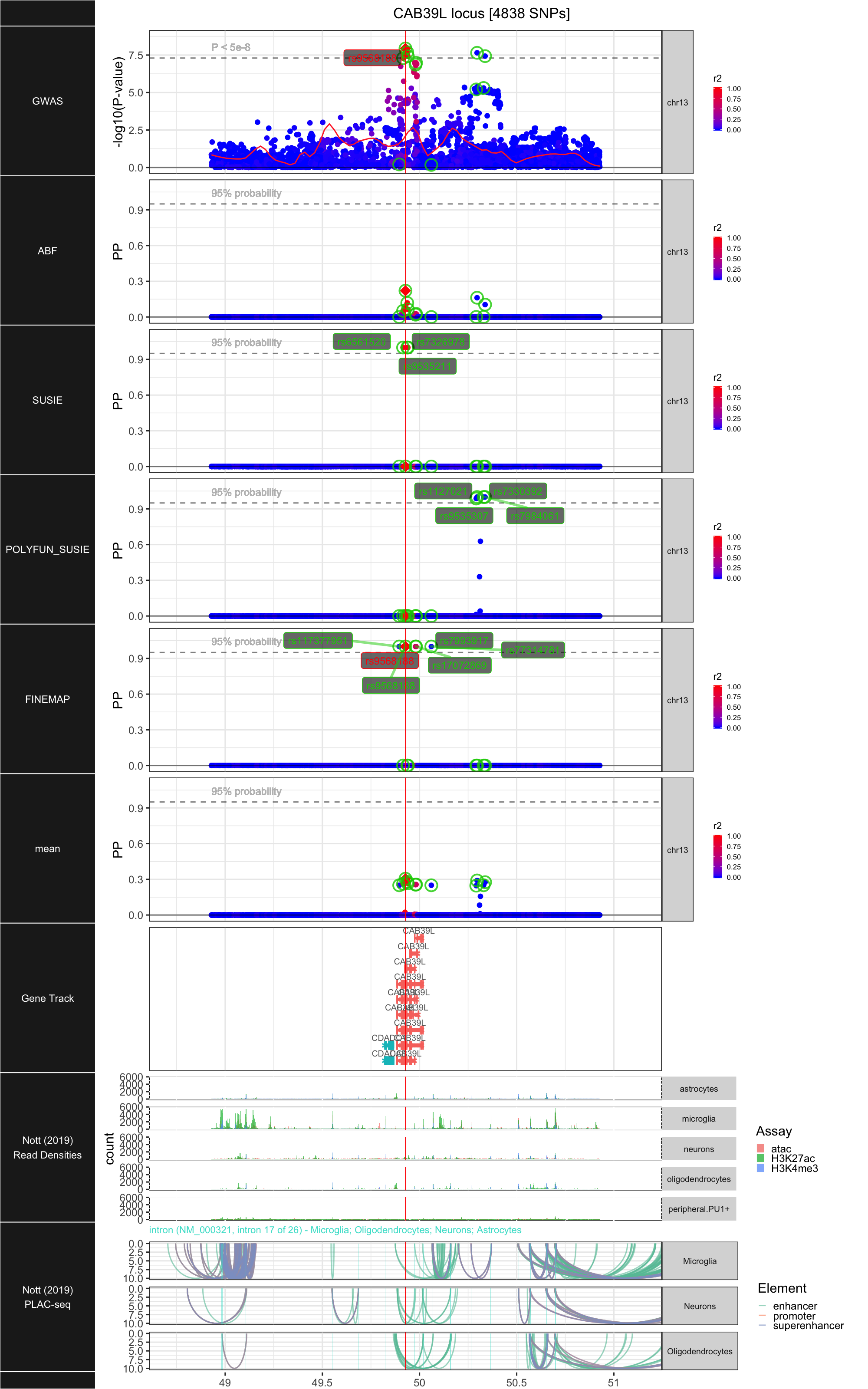
## ### CAB39L
## 
##
## ### CAMK2D
## 
##
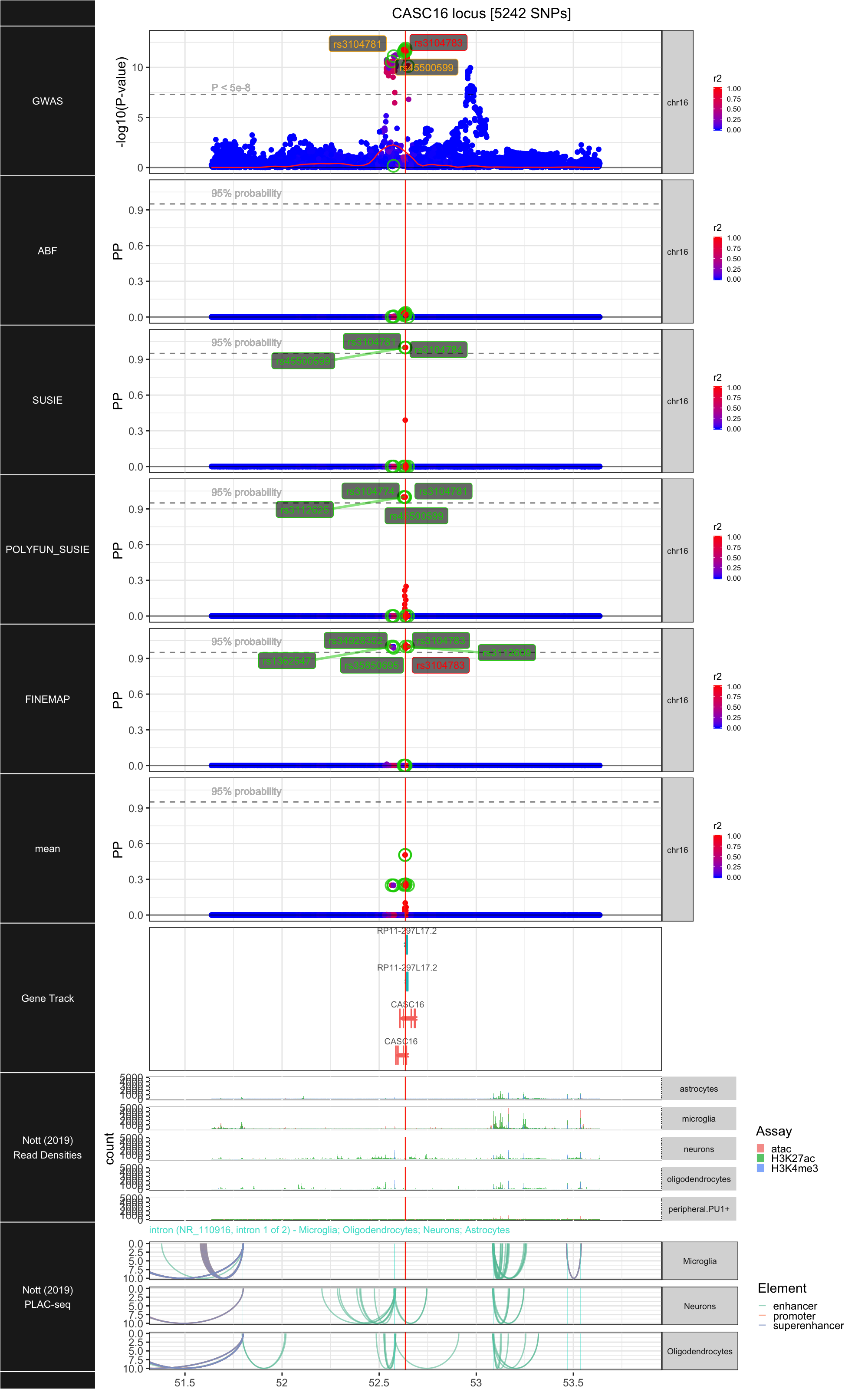
## ### CASC16
## 
##
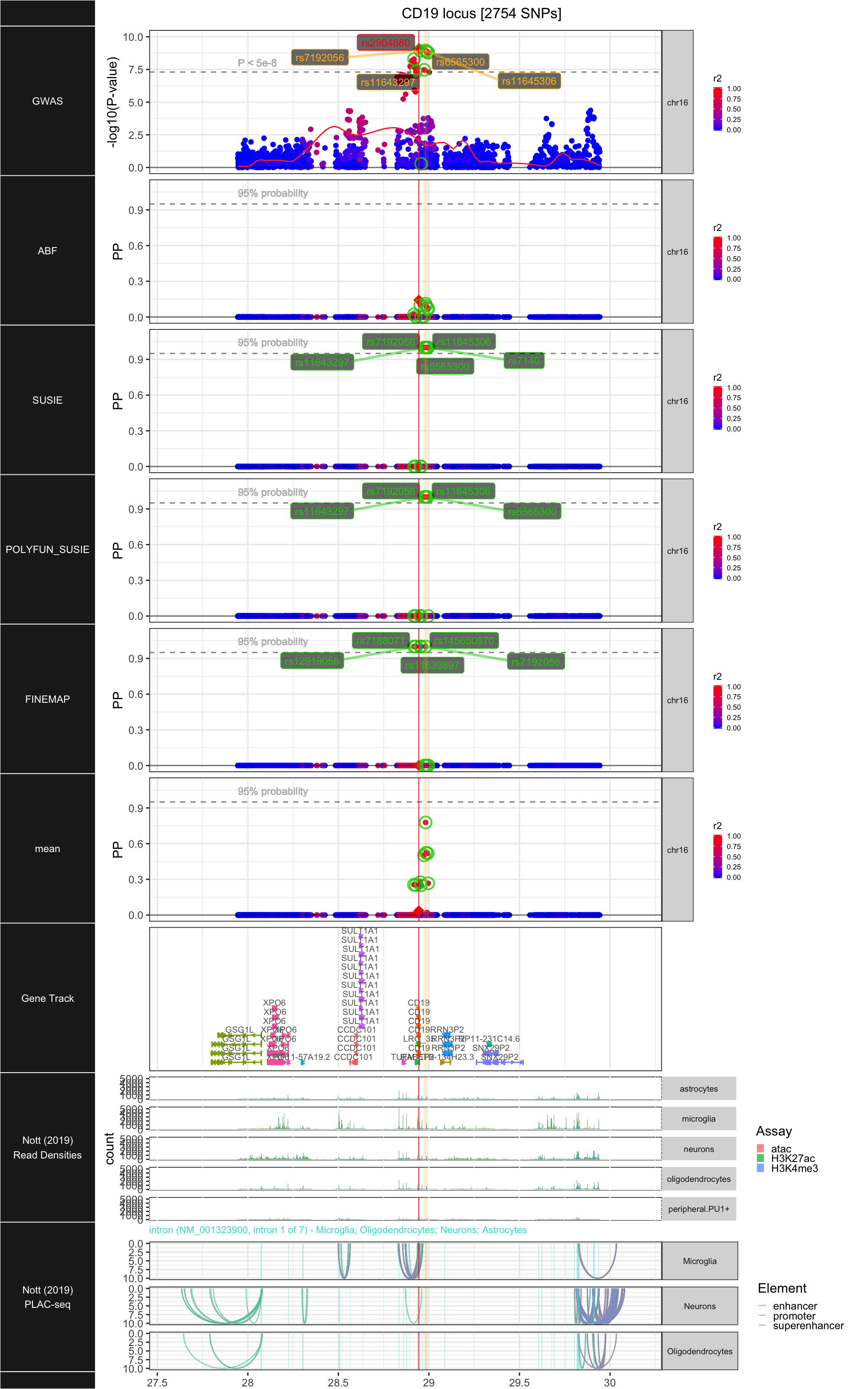
## ### CD19
## 
##
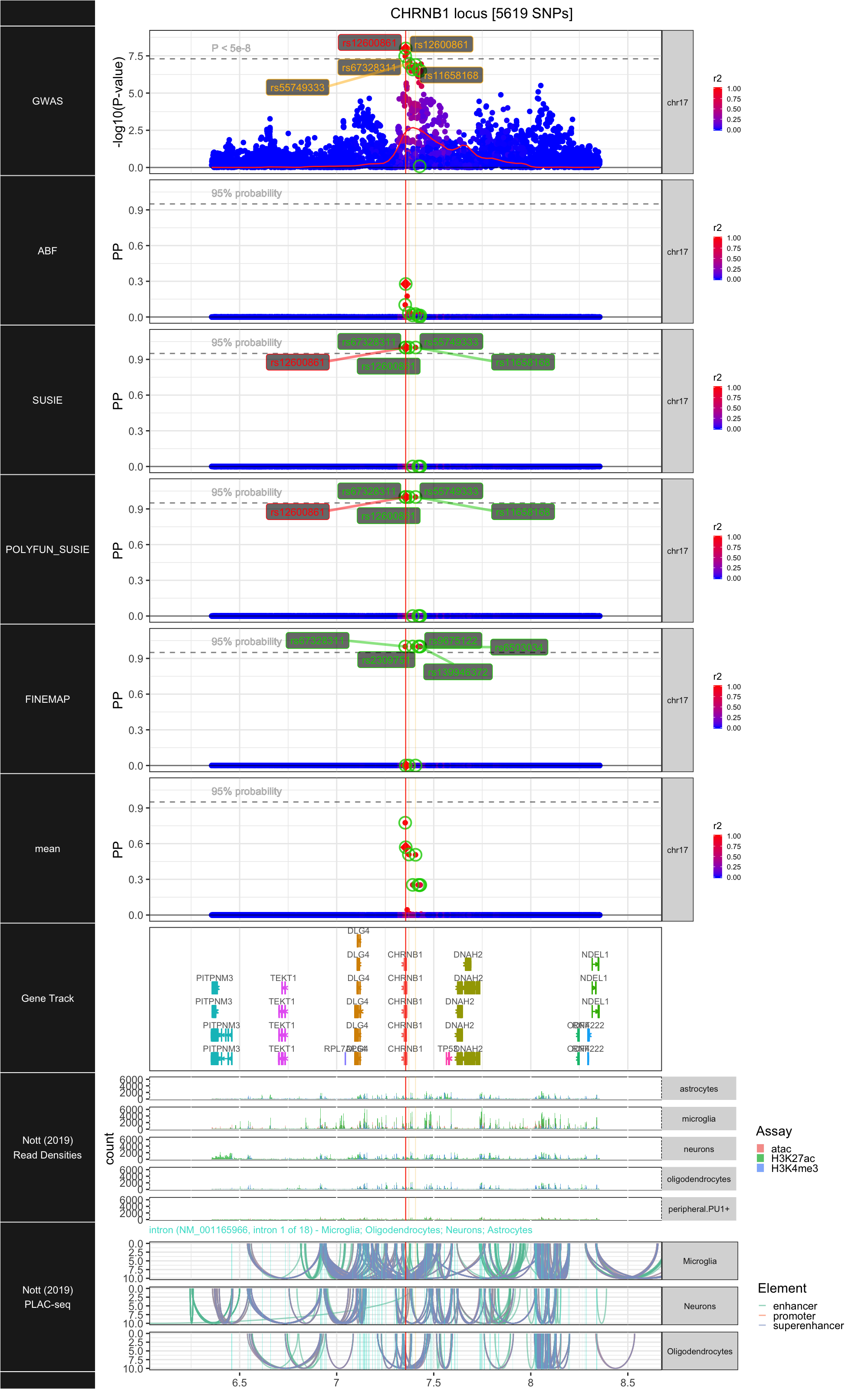
## ### CHRNB1
## 
##
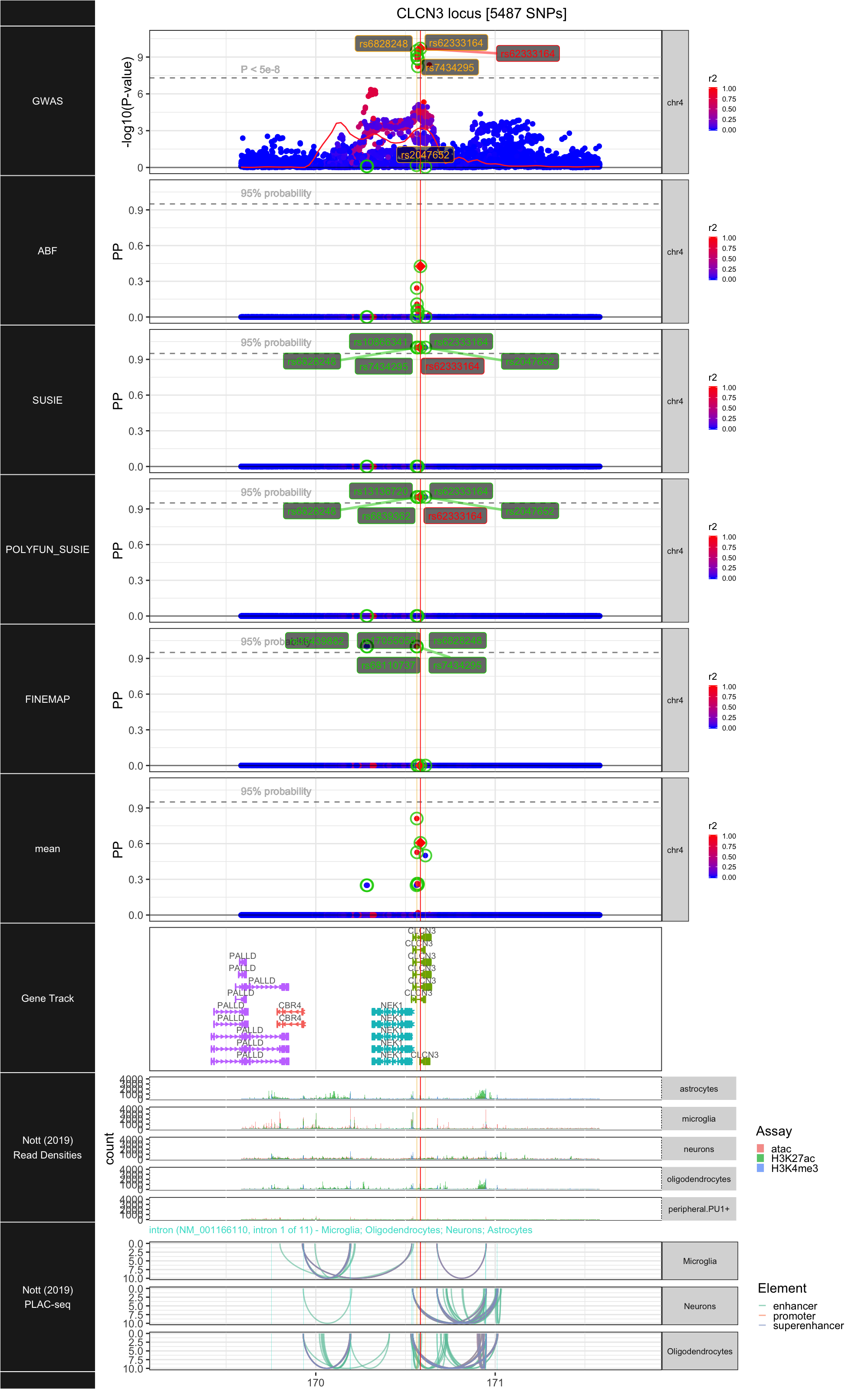
## ### CLCN3
## 
##
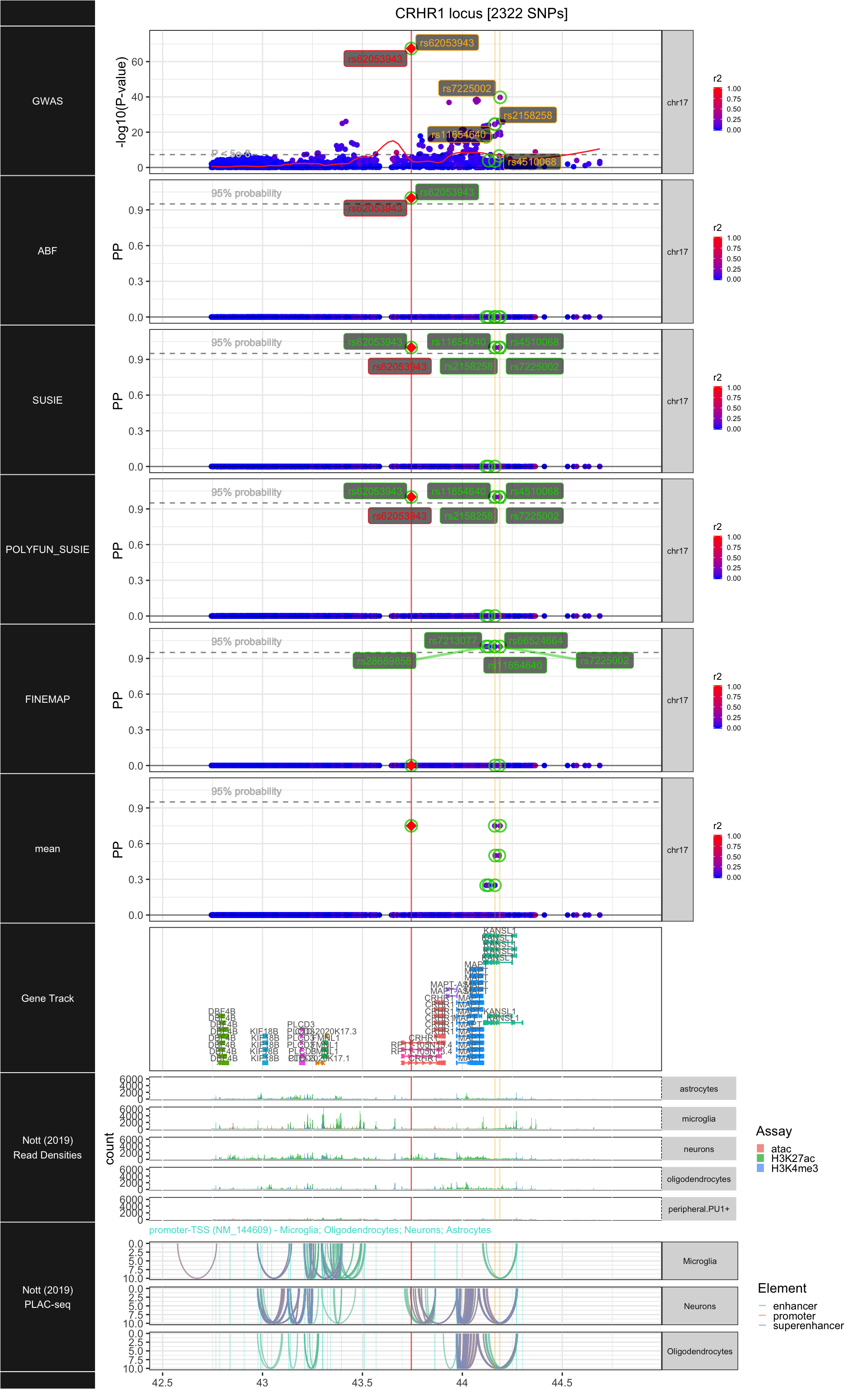
## ### CRHR1
## 
##
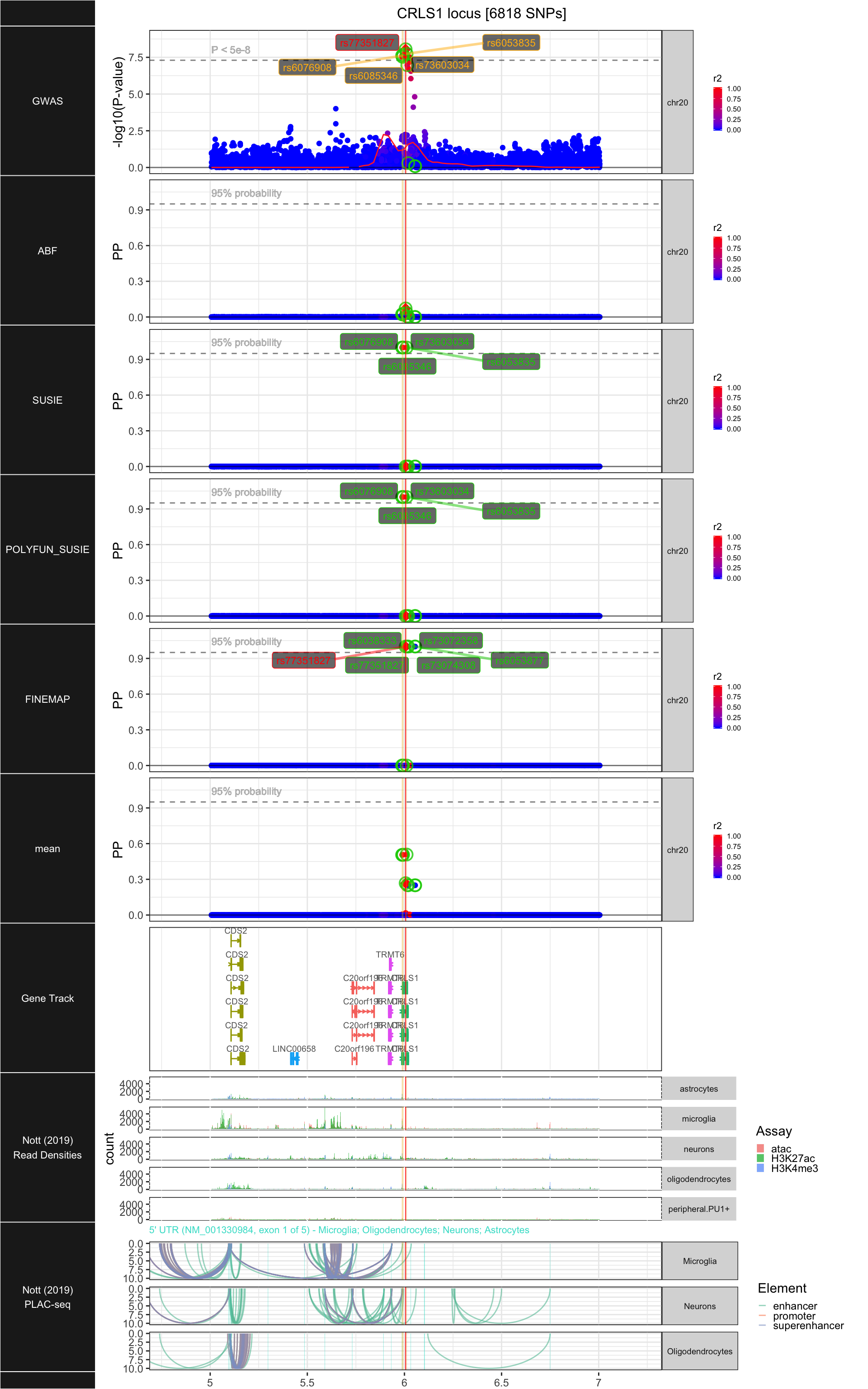
## ### CRLS1
## 
##
## ### CTSB
## 
##
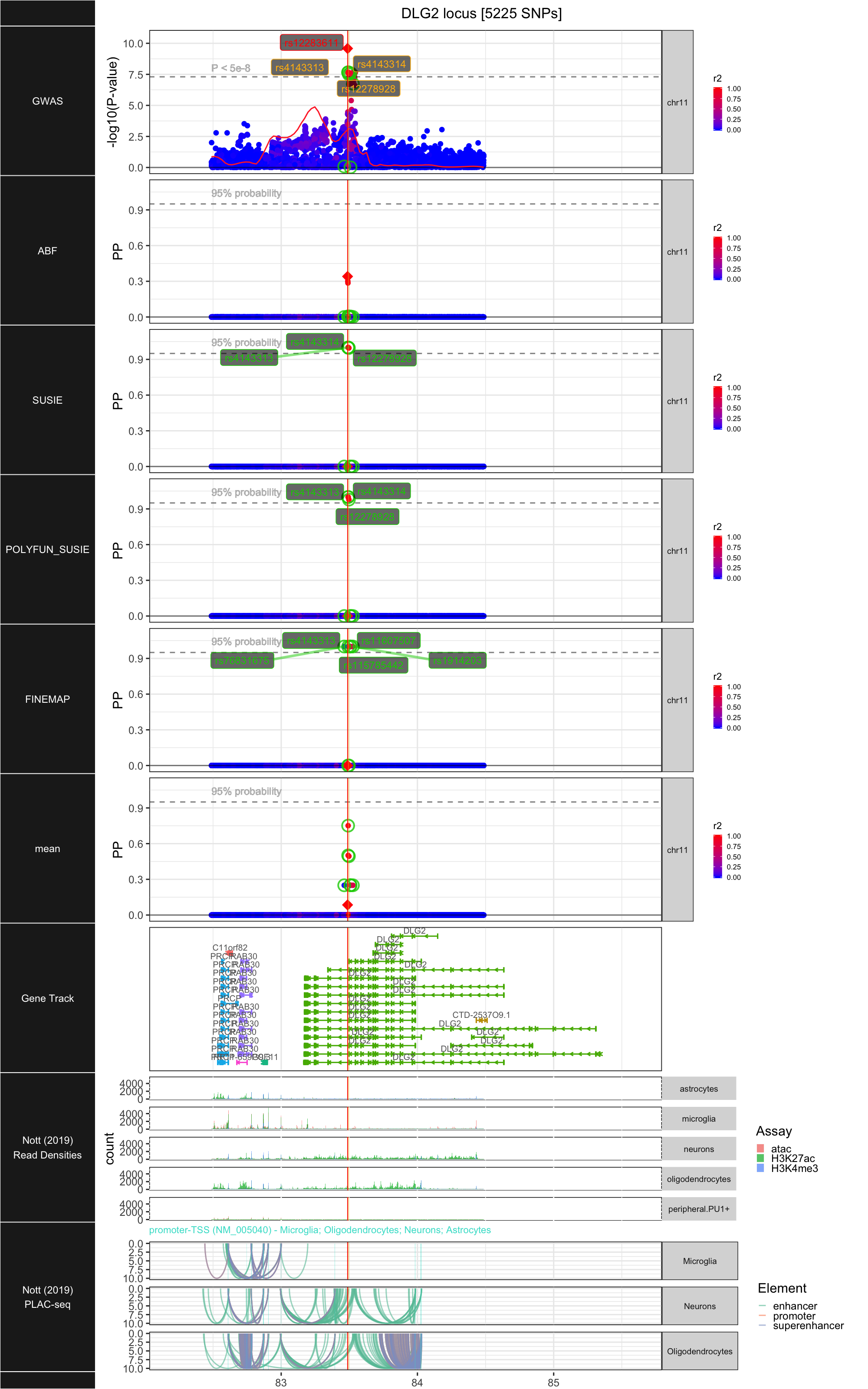
## ### DLG2
## 
##
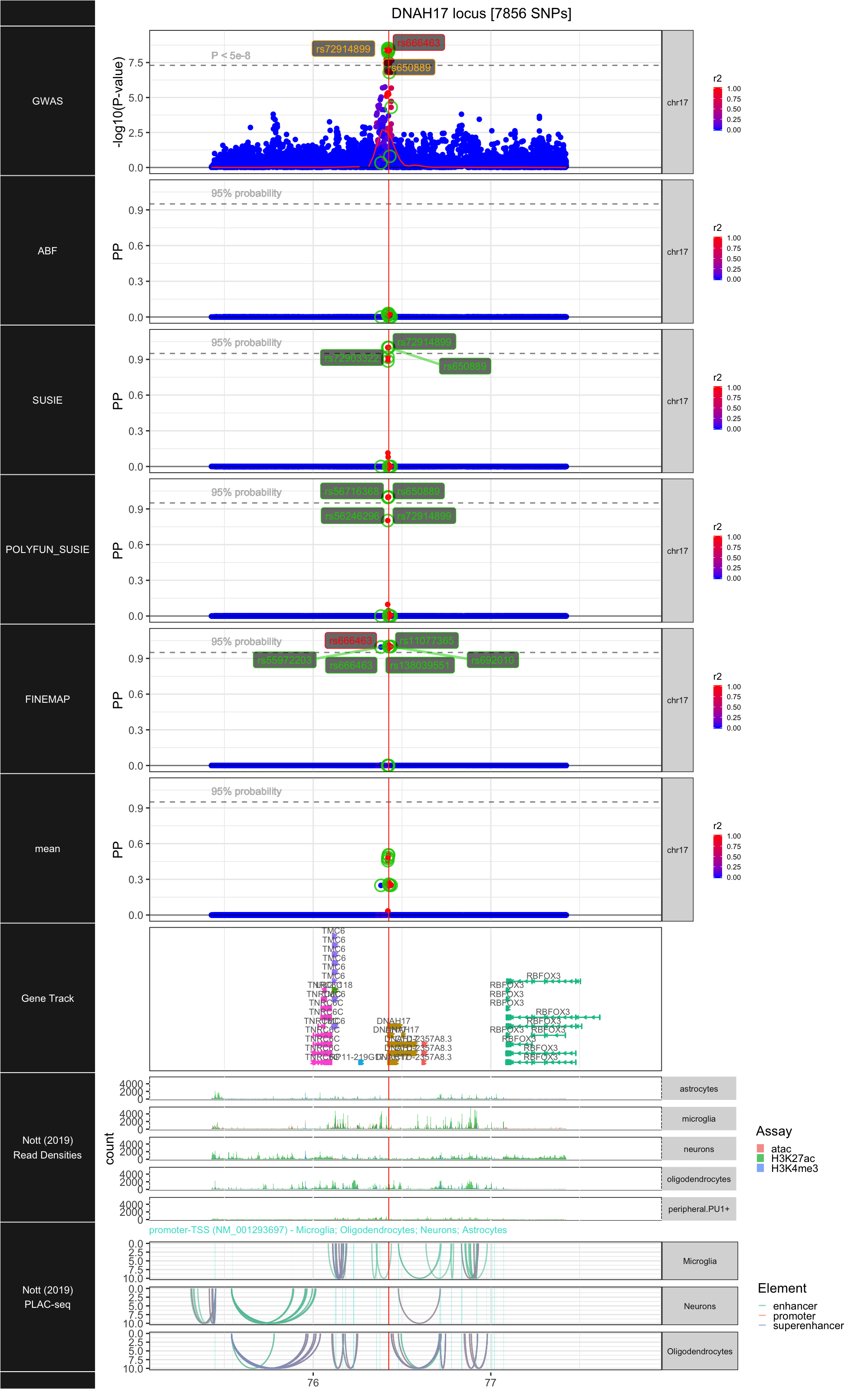
## ### DNAH17
## 
##
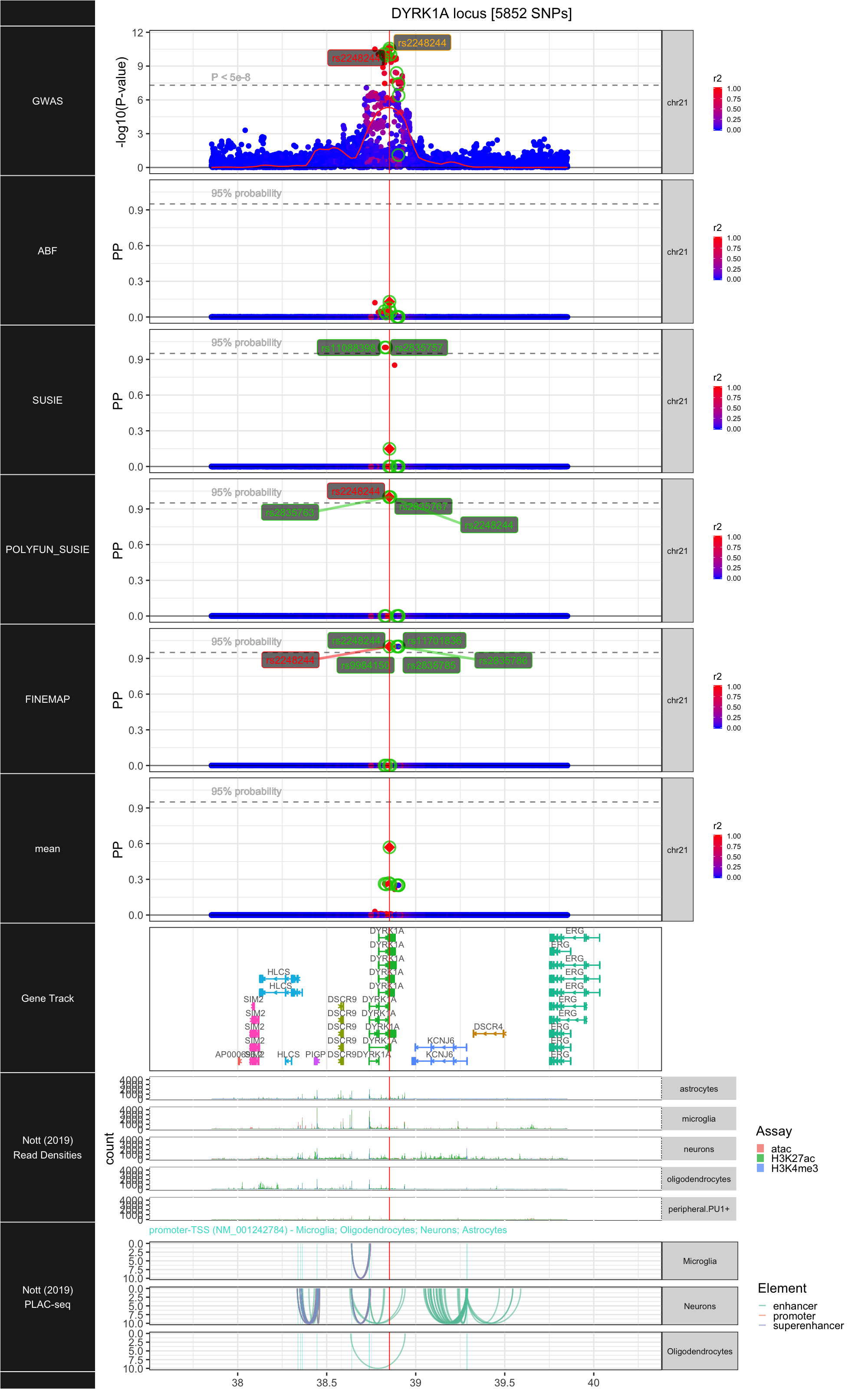
## ### DYRK1A
## 
##
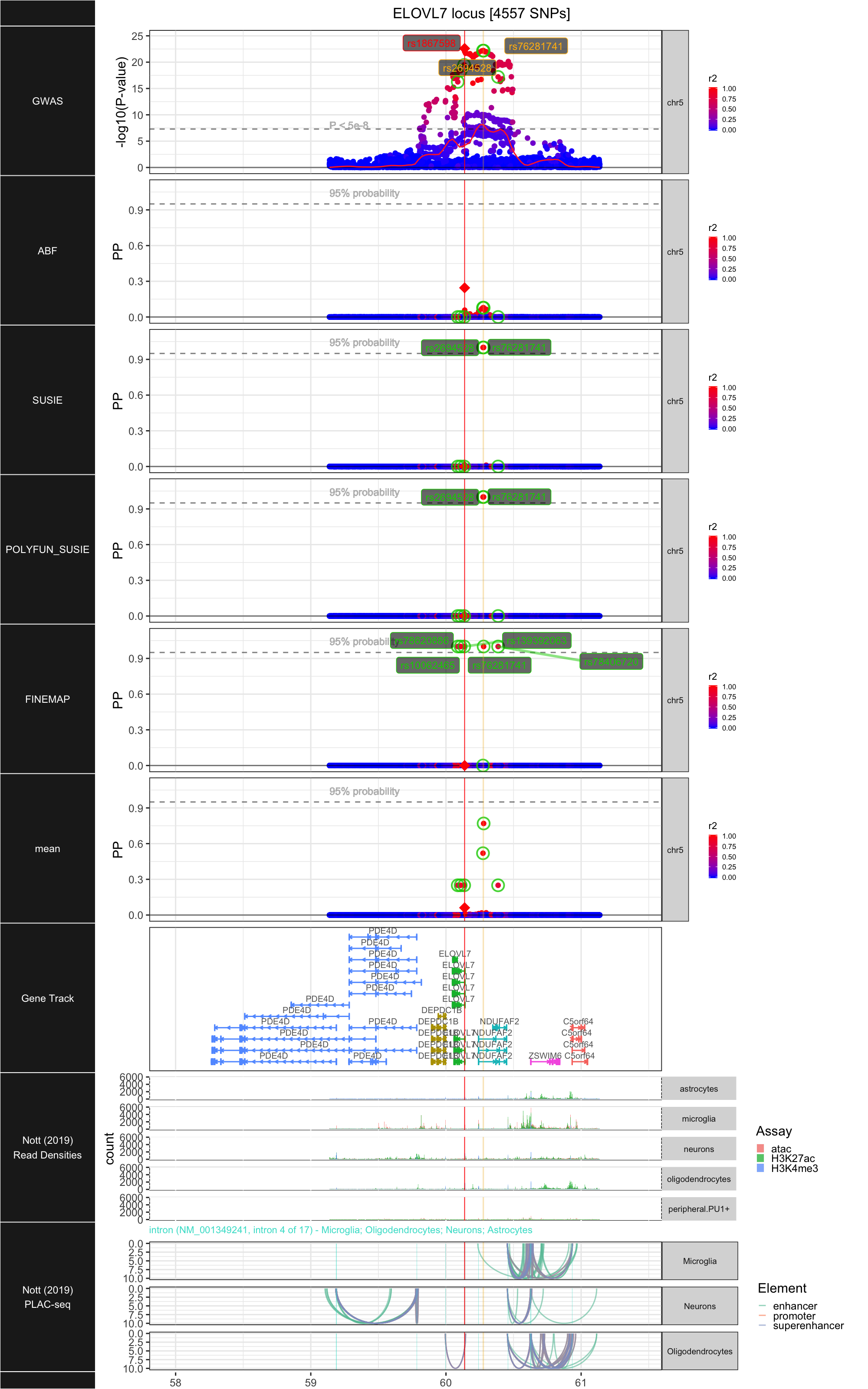
## ### ELOVL7
## 
##
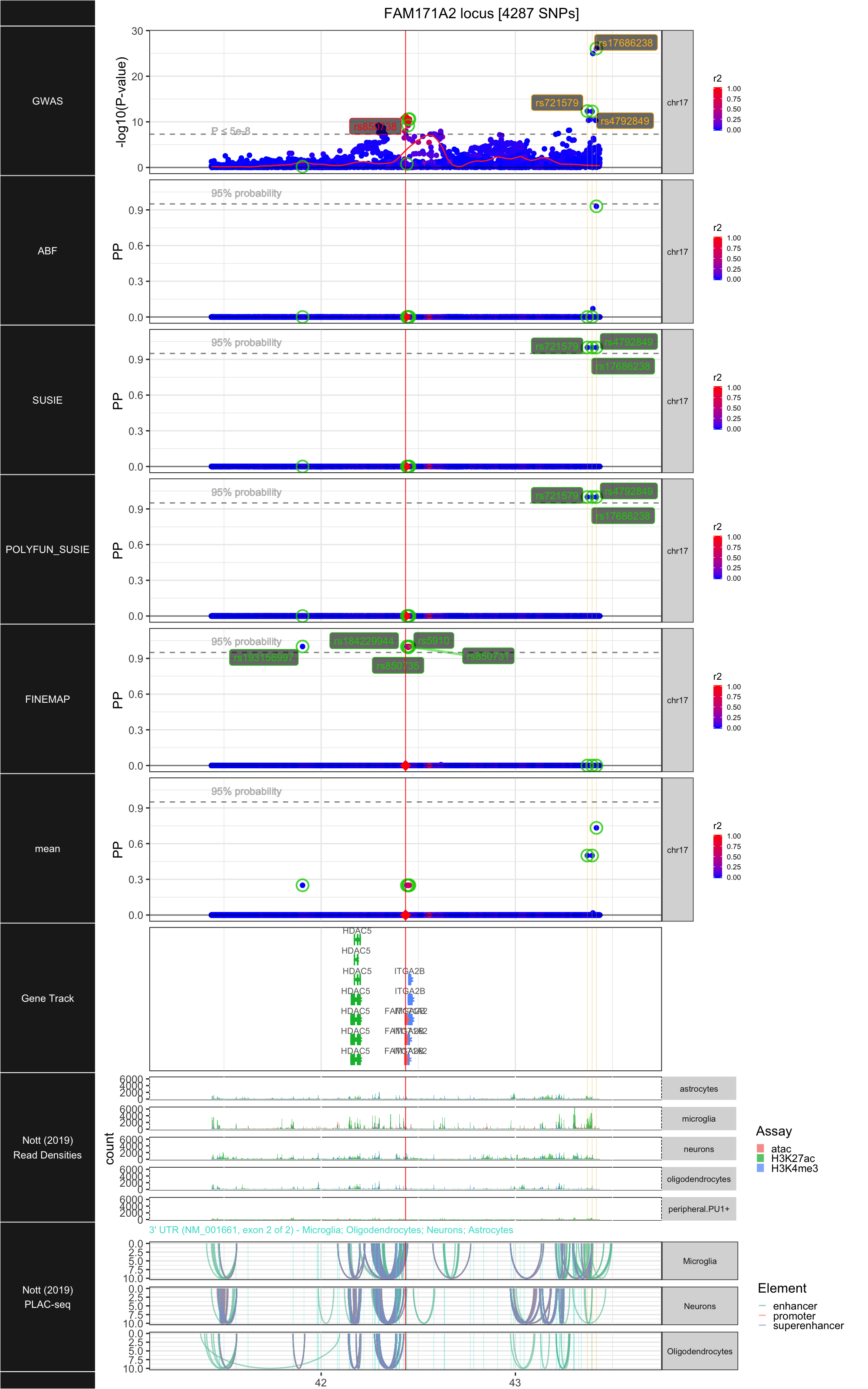
## ### FAM171A2
## 
##
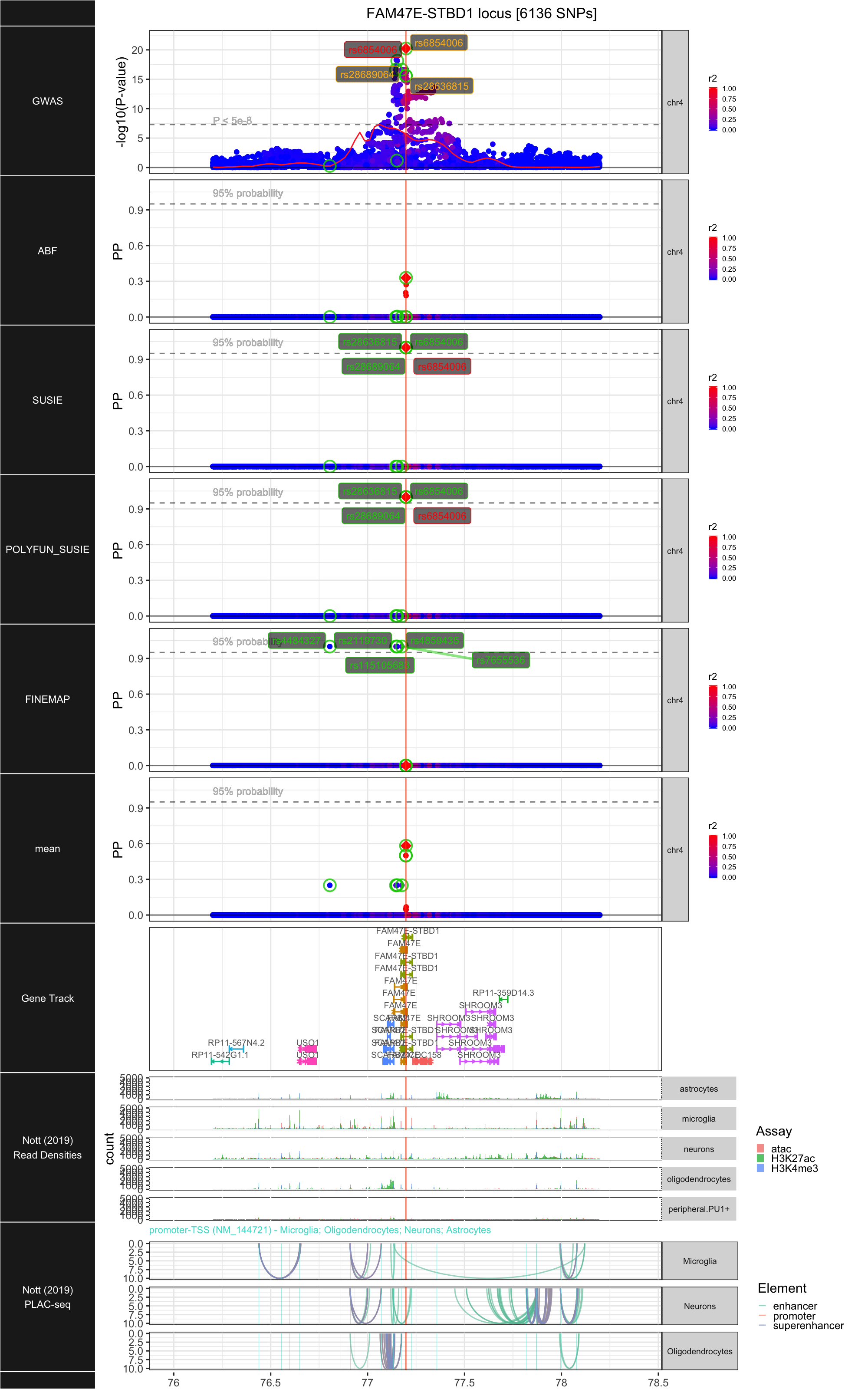
## ### FAM47E-STBD1
## 
##
## ### FAM49B
## 
##
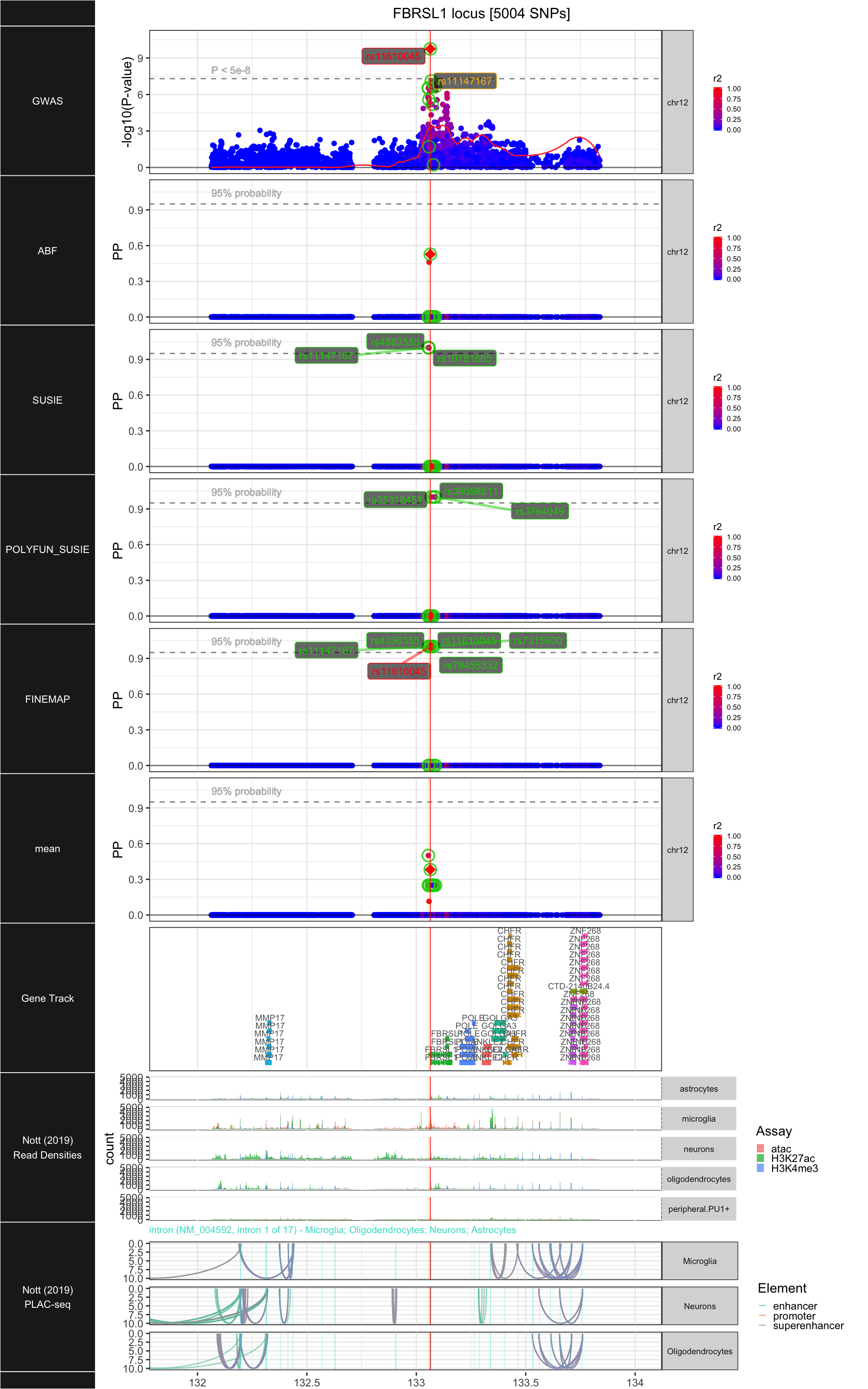
## ### FBRSL1
## 
##
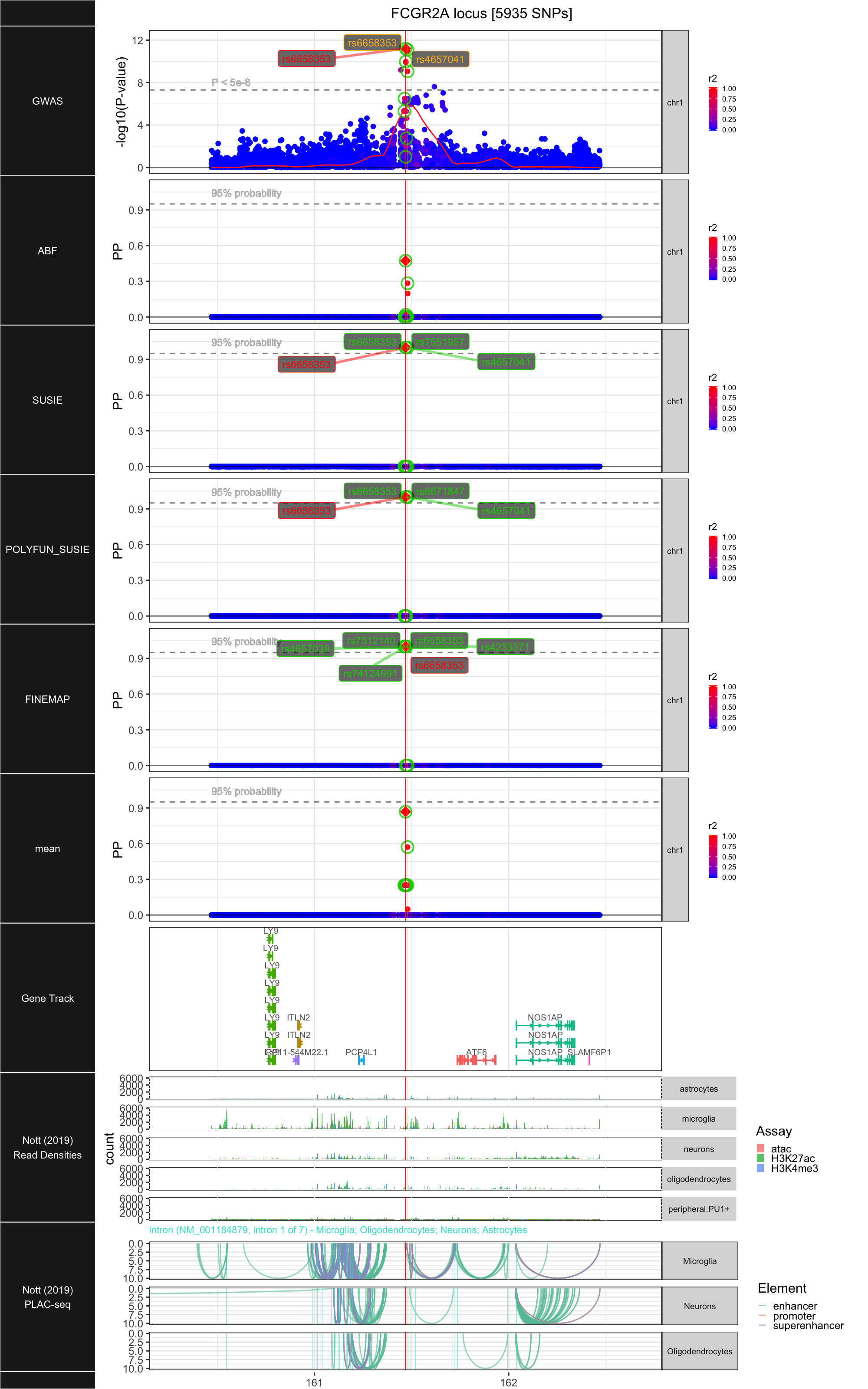
## ### FCGR2A
## 
##
## ### FGF20
## 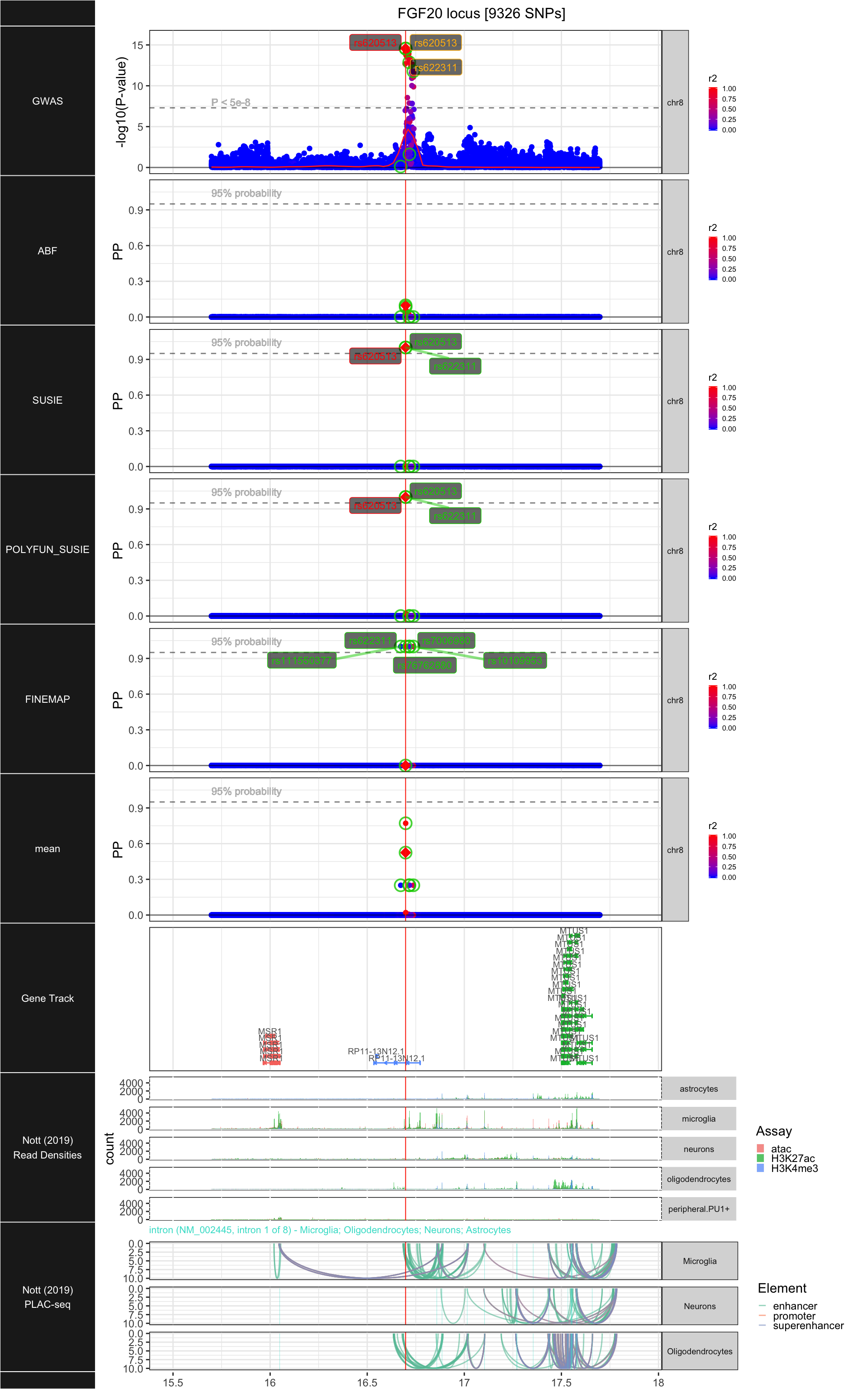
##
## ### FYN
## 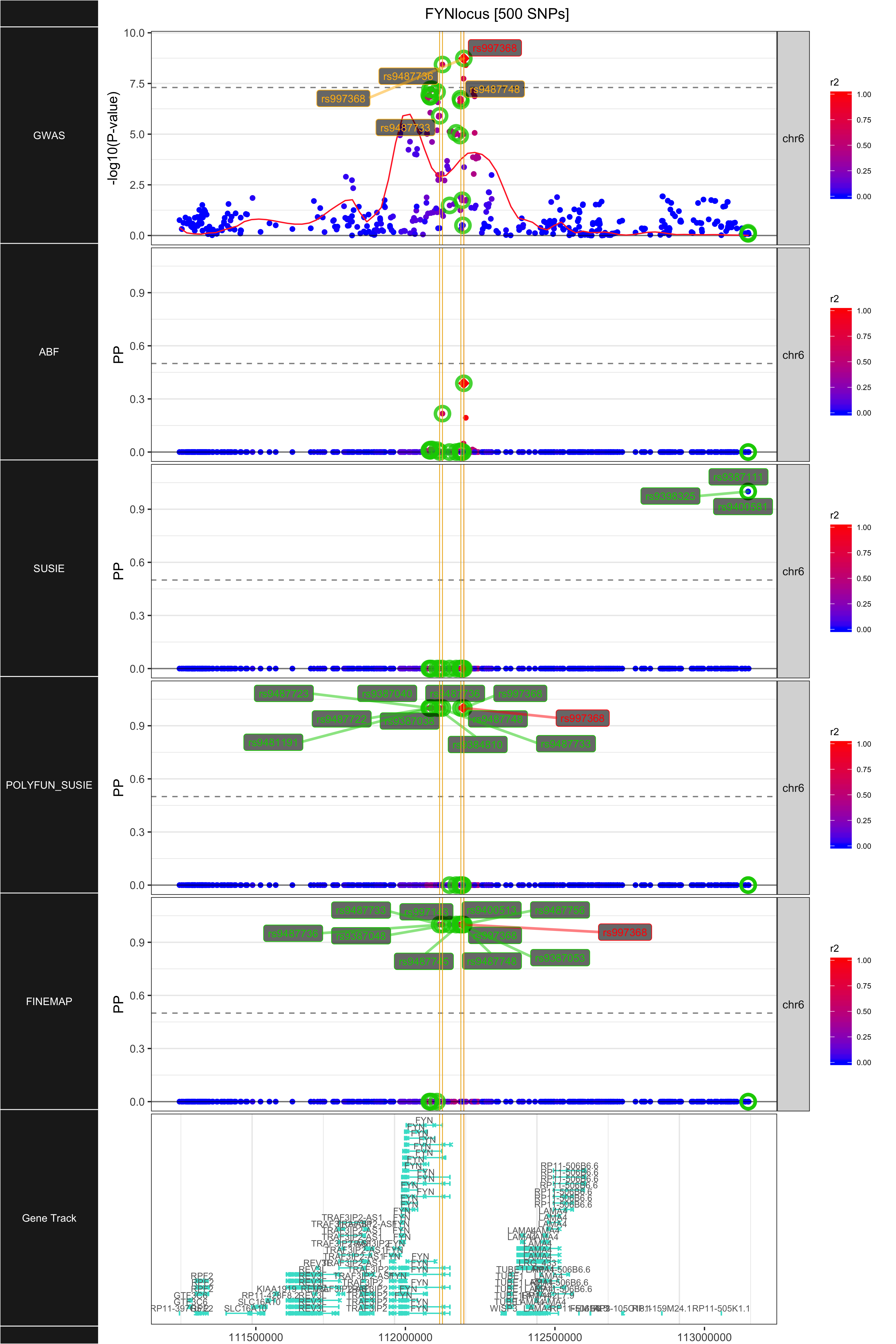
##
## ### GALC
## 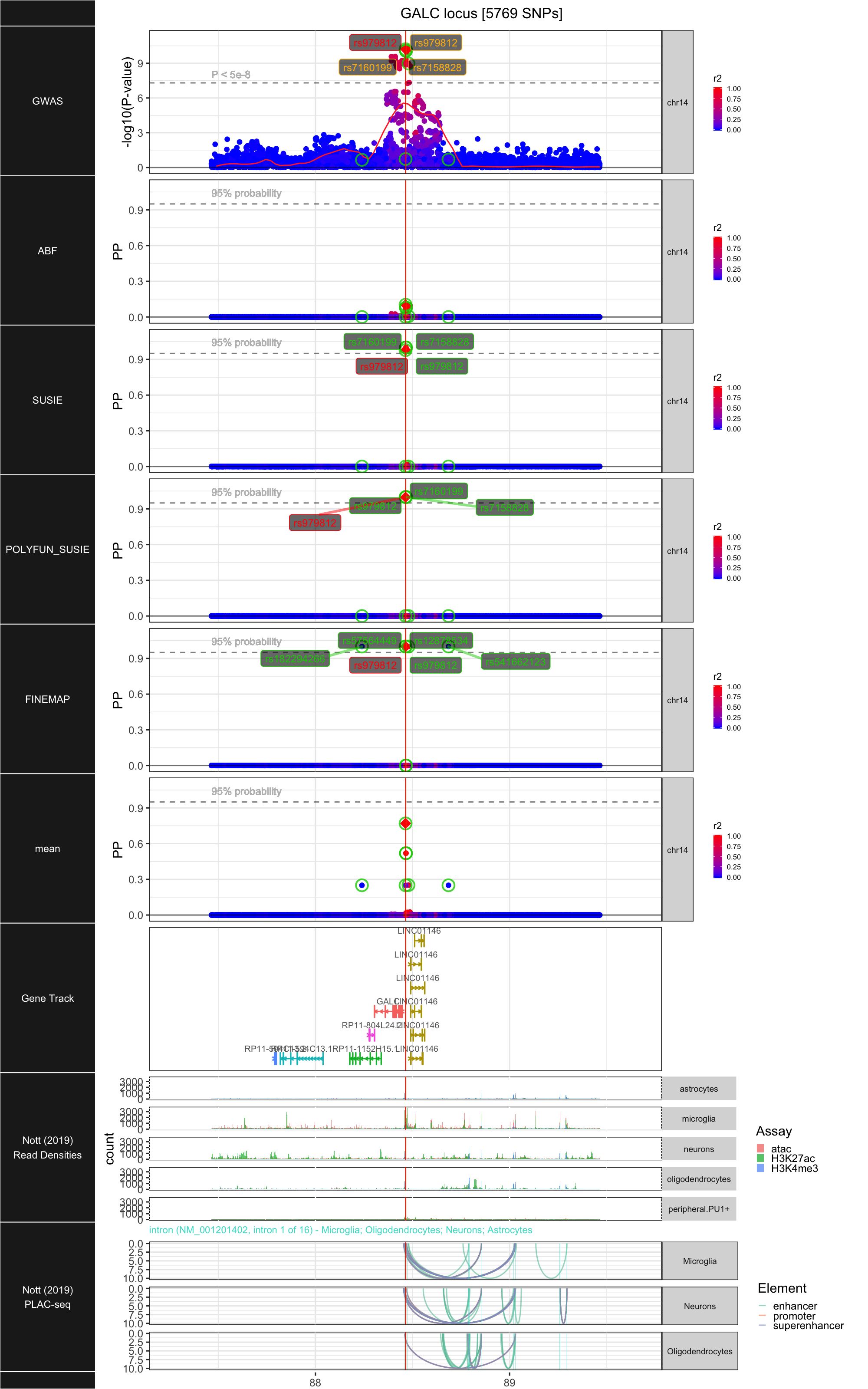
##
## ### GBF1
## 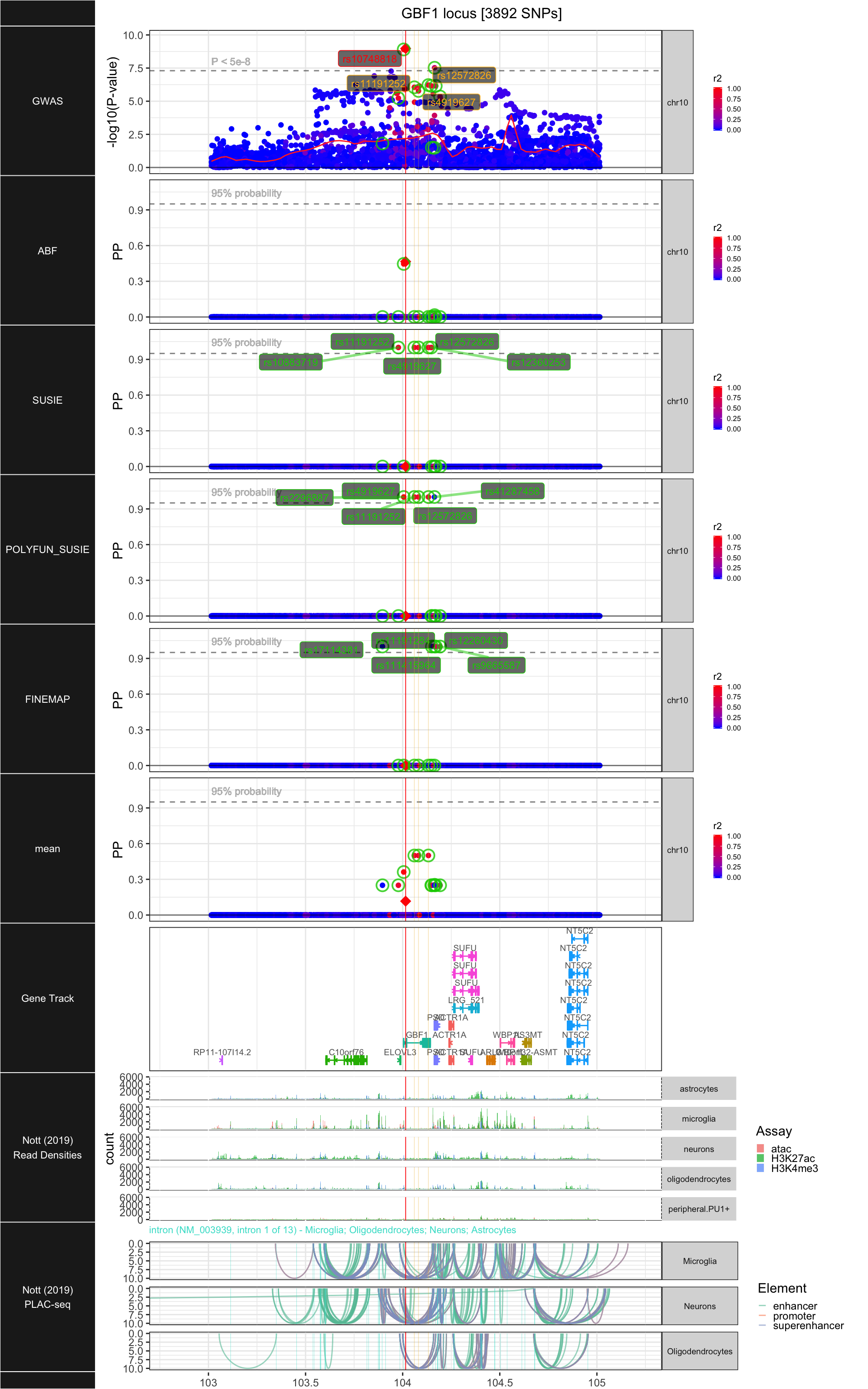
##
## ### GCH1
## 
##
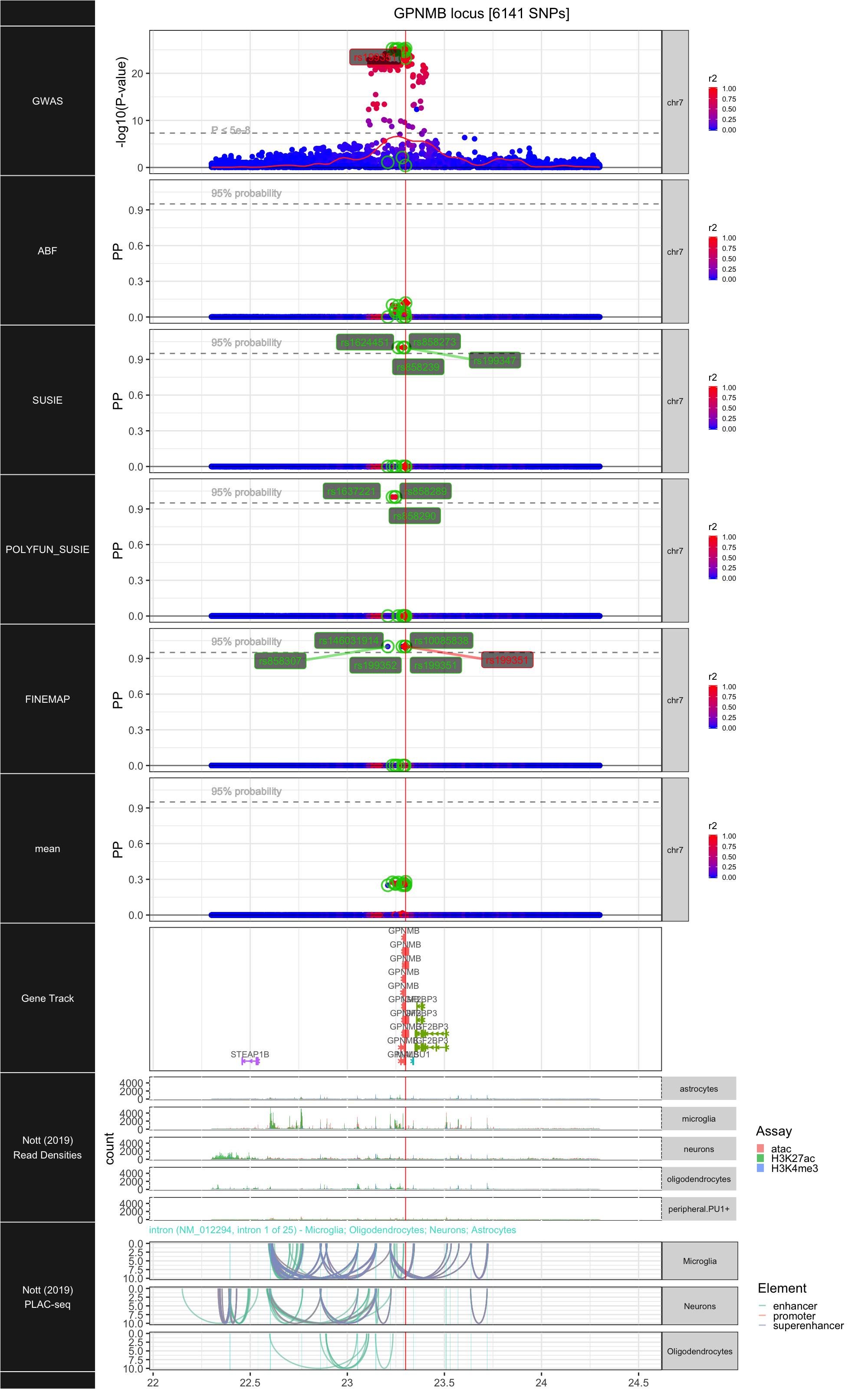
## ### GPNMB
## 
##
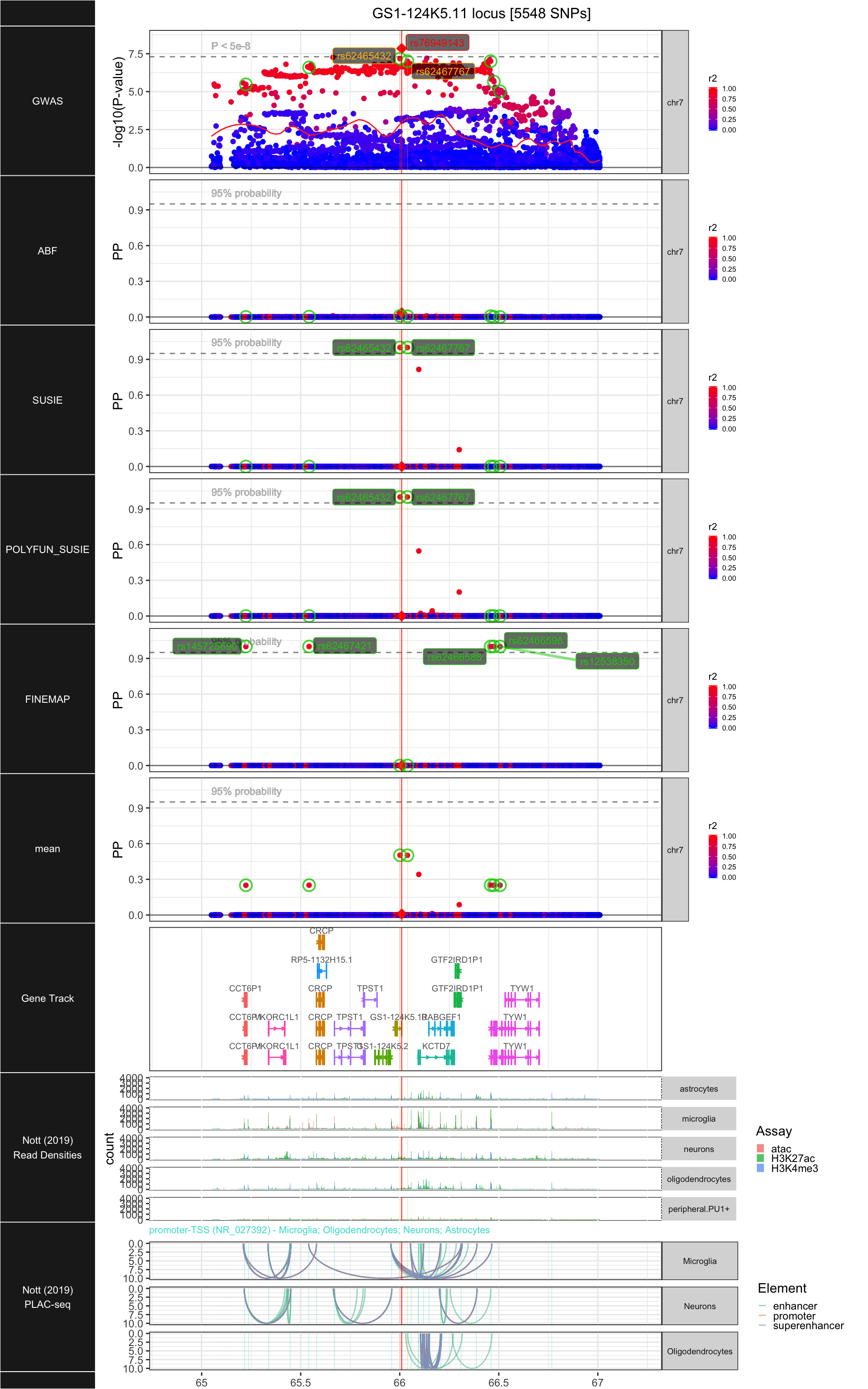
## ### GS1-124K5.11
## 
##
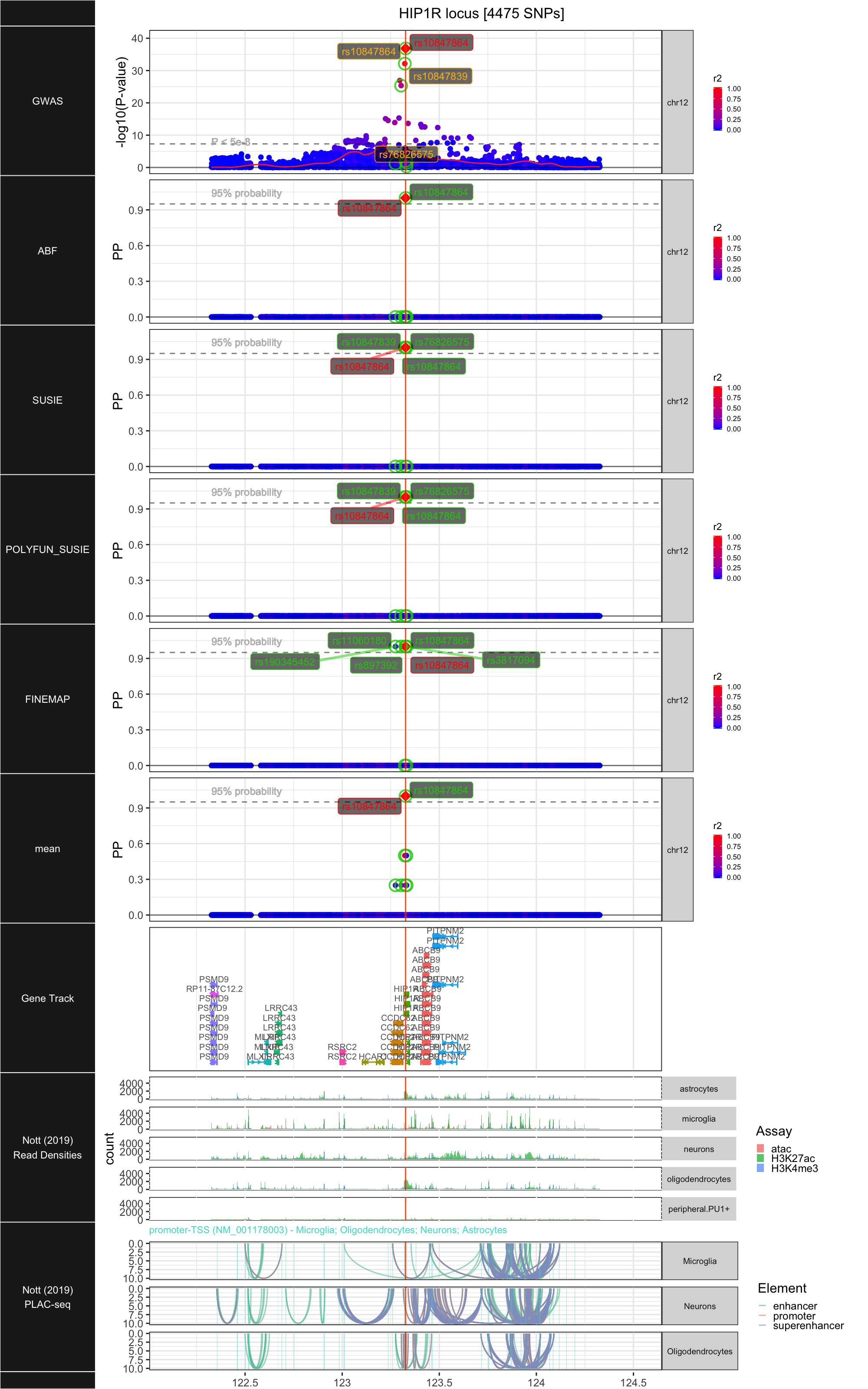
## ### HIP1R
## 
##
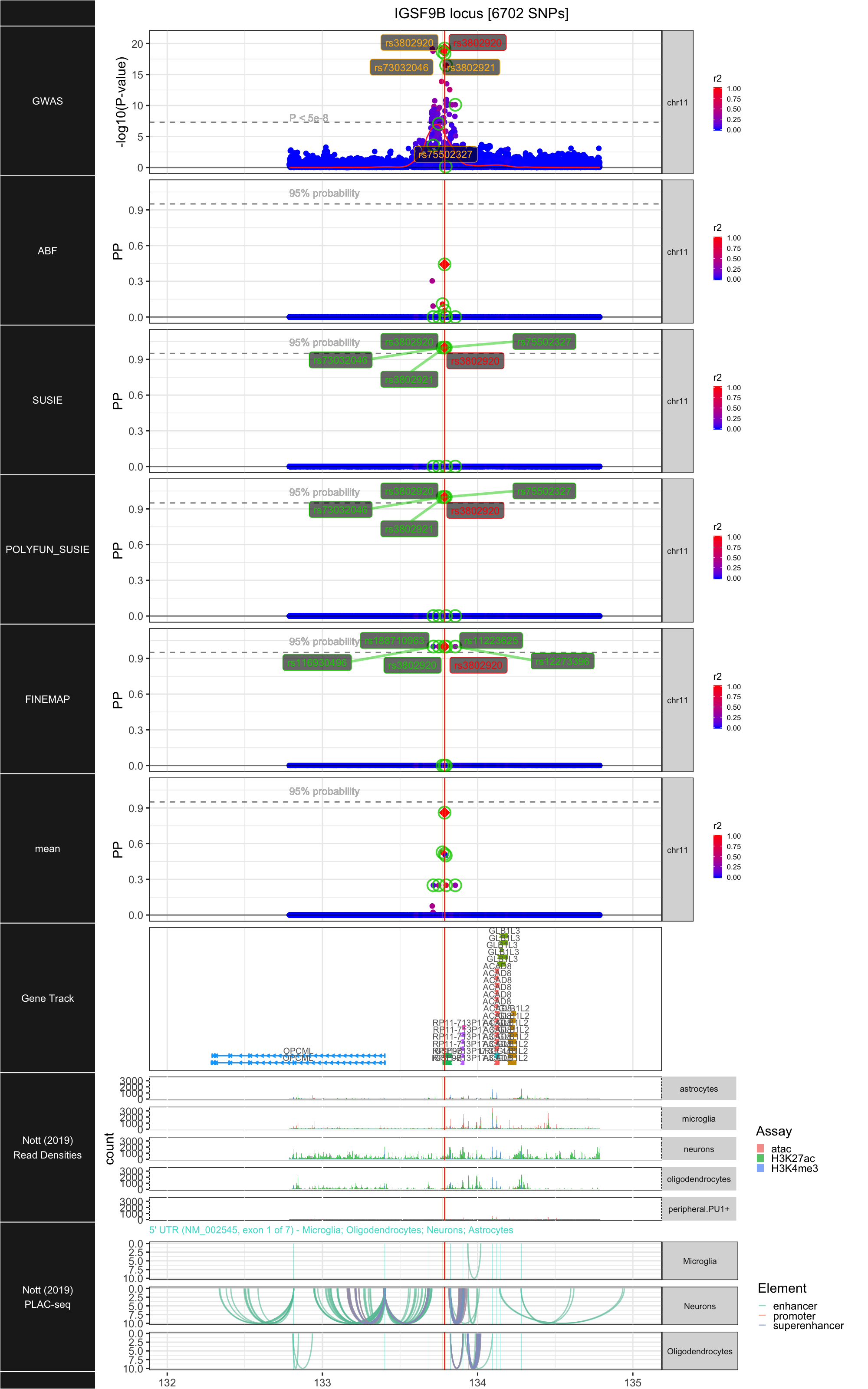
## ### IGSF9B
## 
##
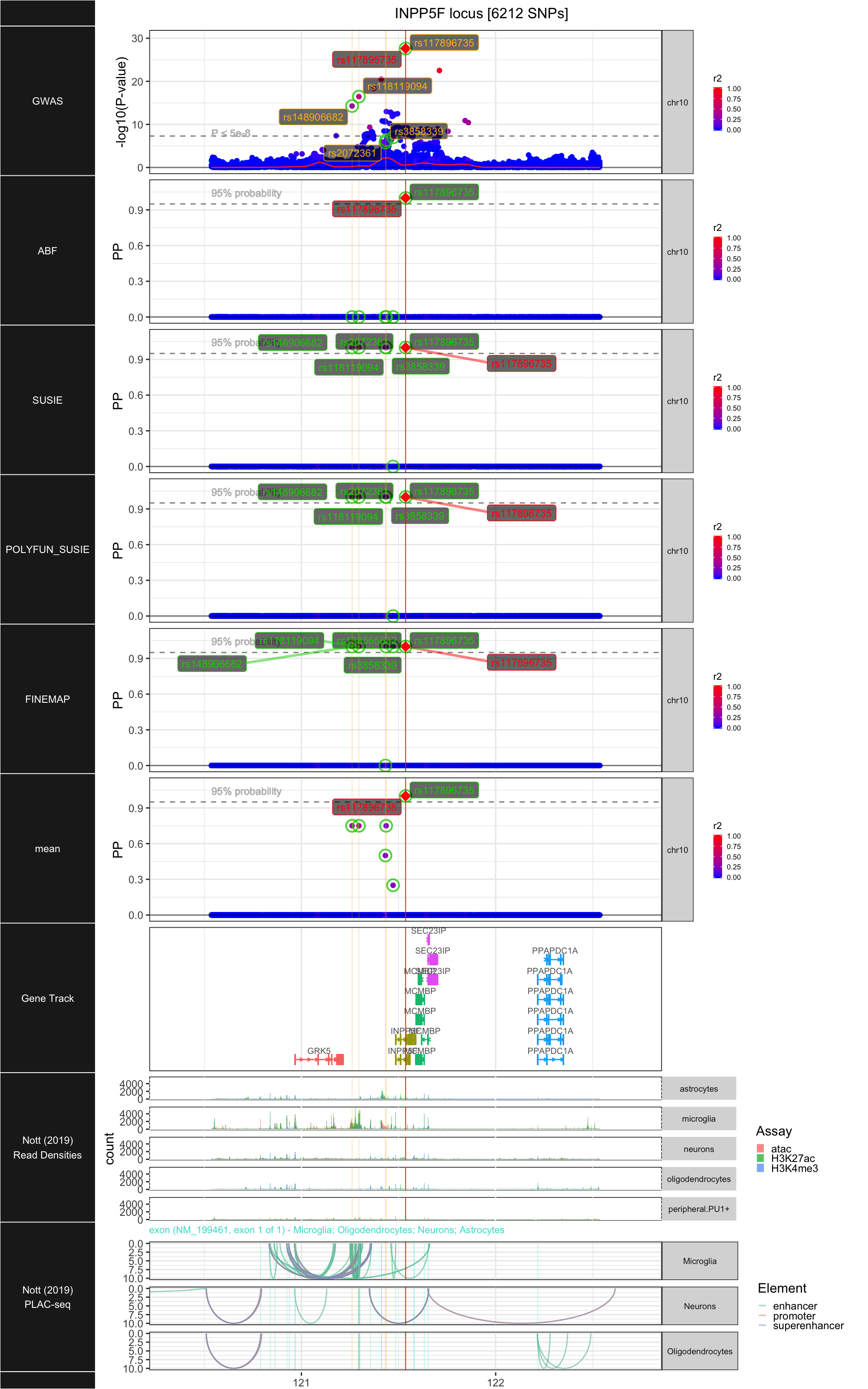
## ### INPP5F
## 
##
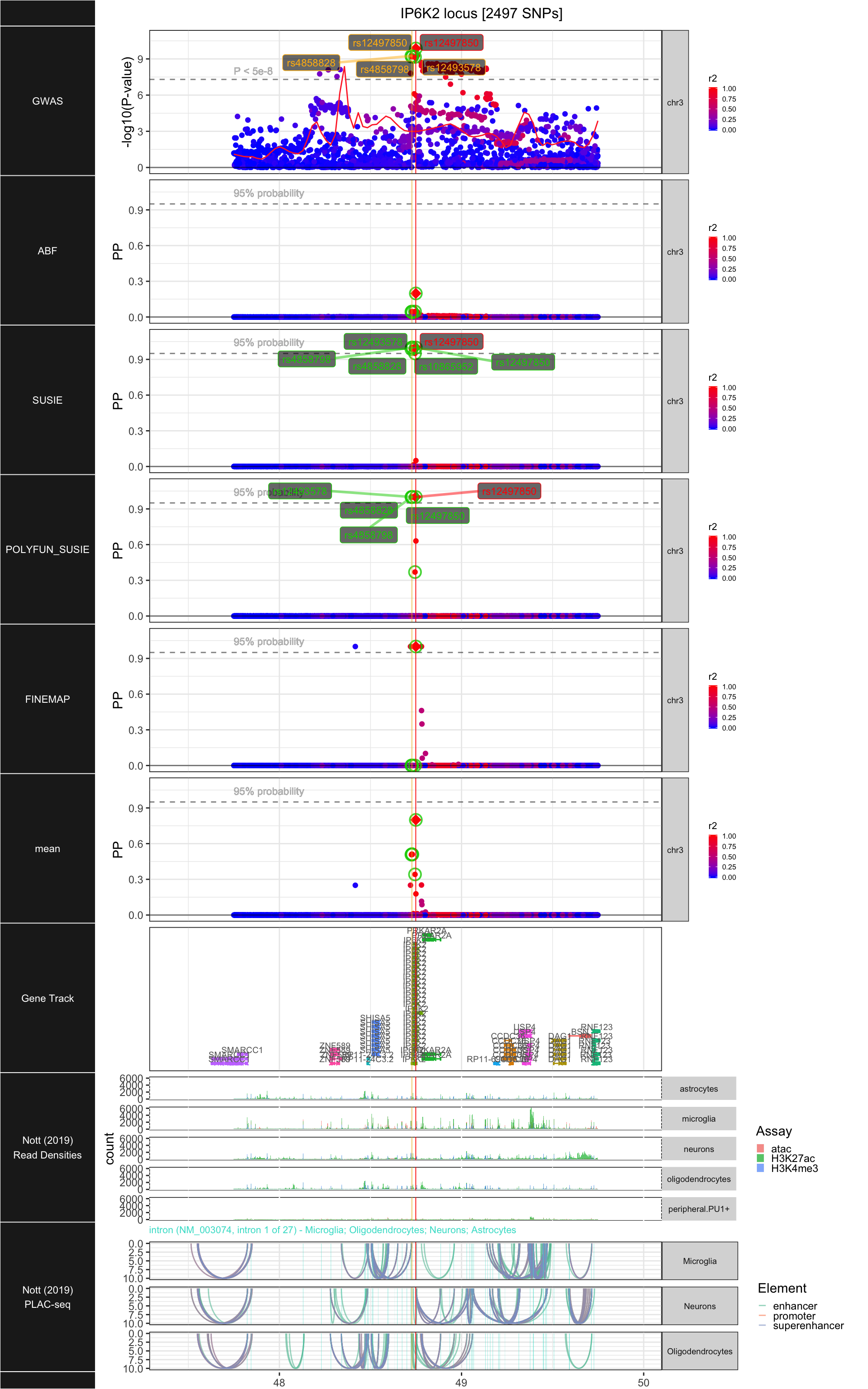
## ### IP6K2
## 
##
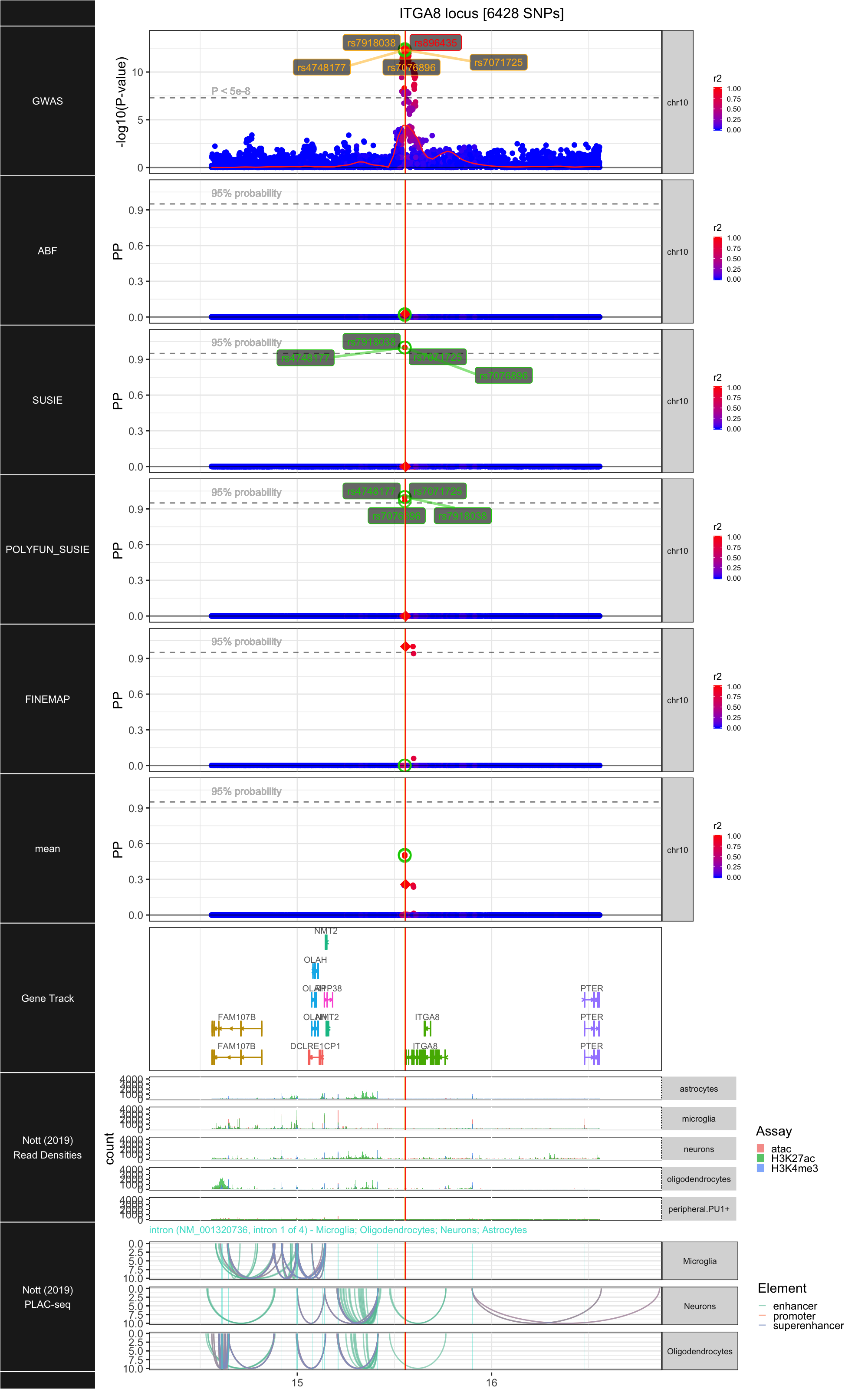
## ### ITGA8
## 
##
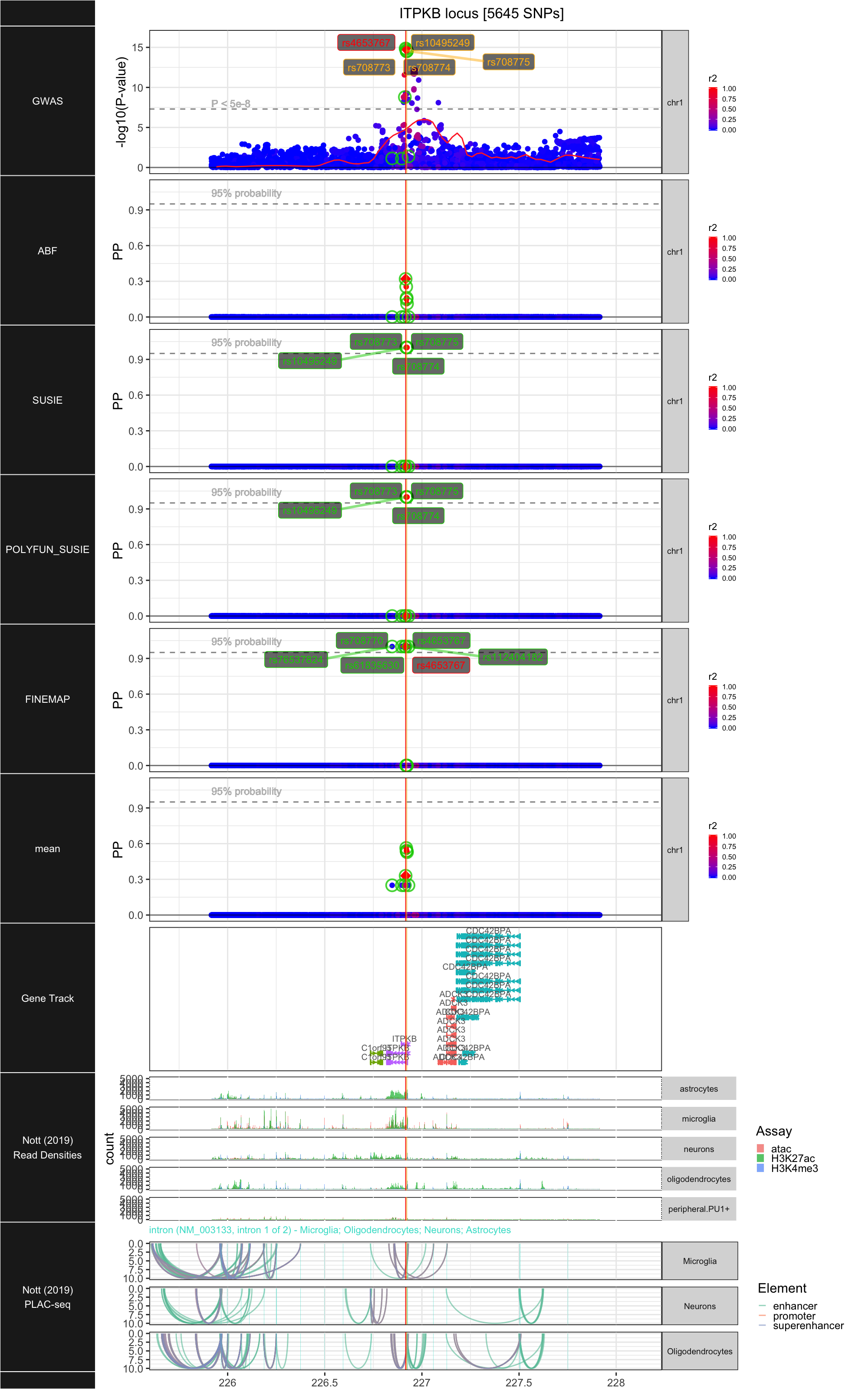
## ### ITPKB
## 
##
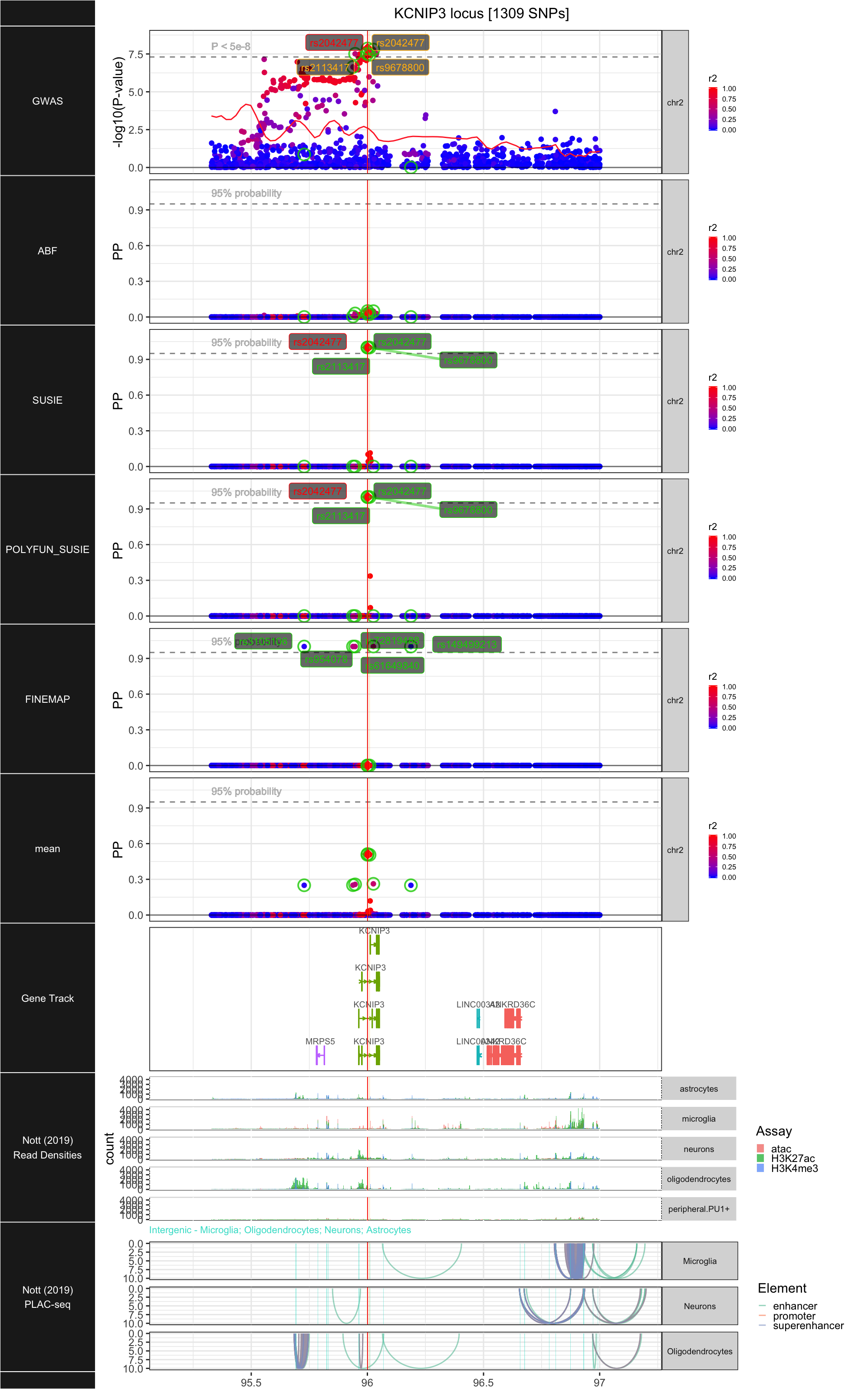
## ### KCNIP3
## 
##
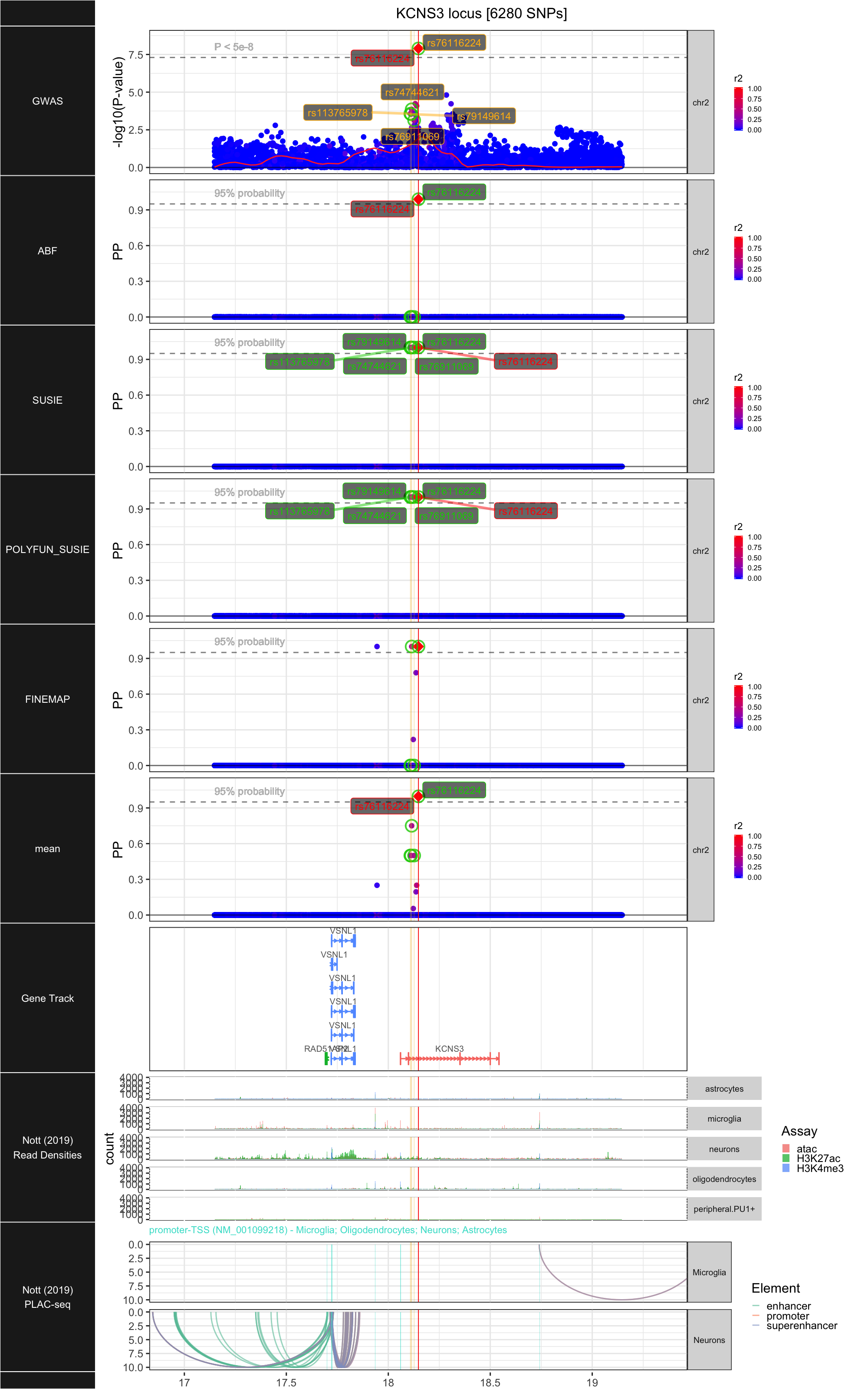
## ### KCNS3
## 
##
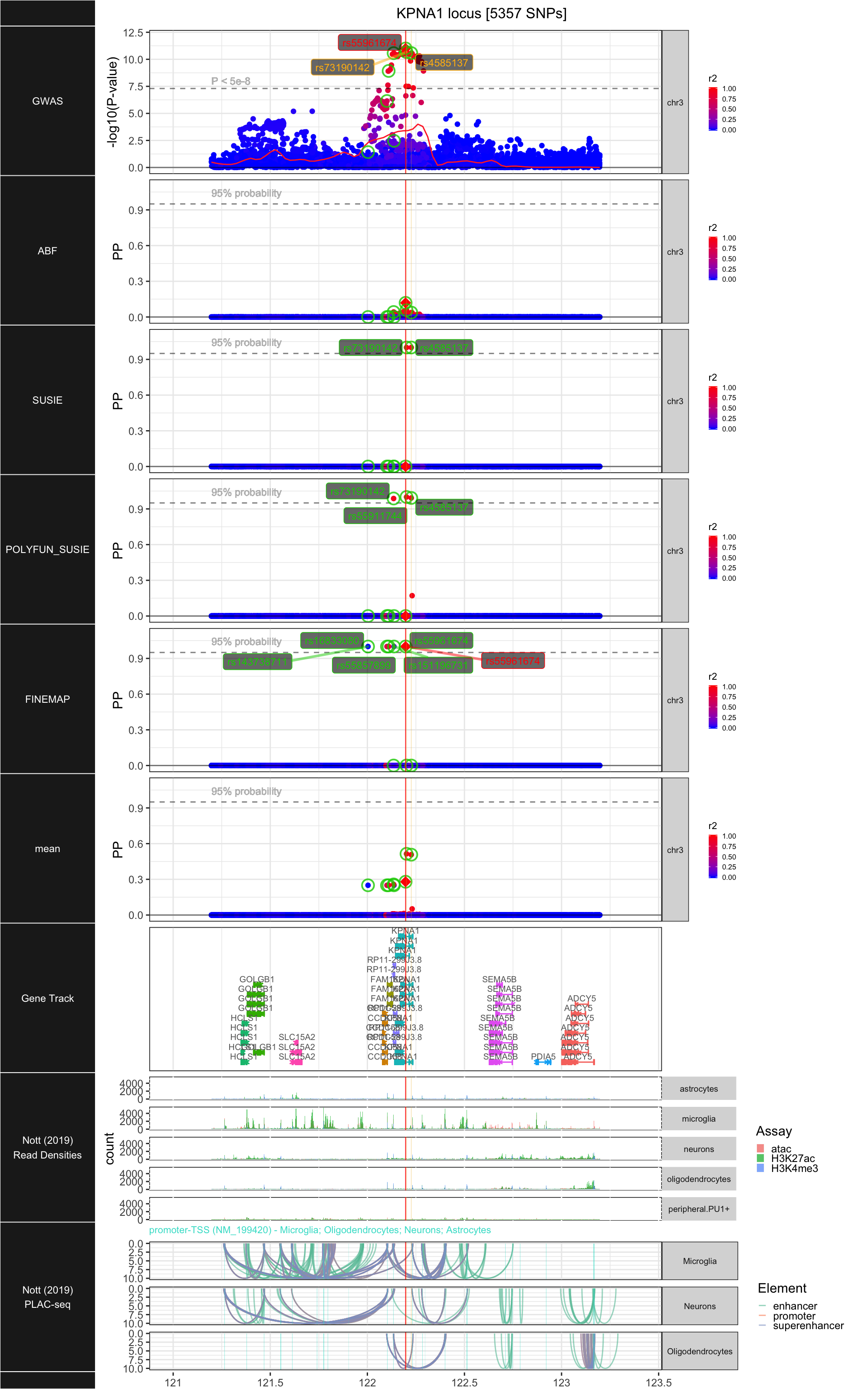
## ### KPNA1
## 
##
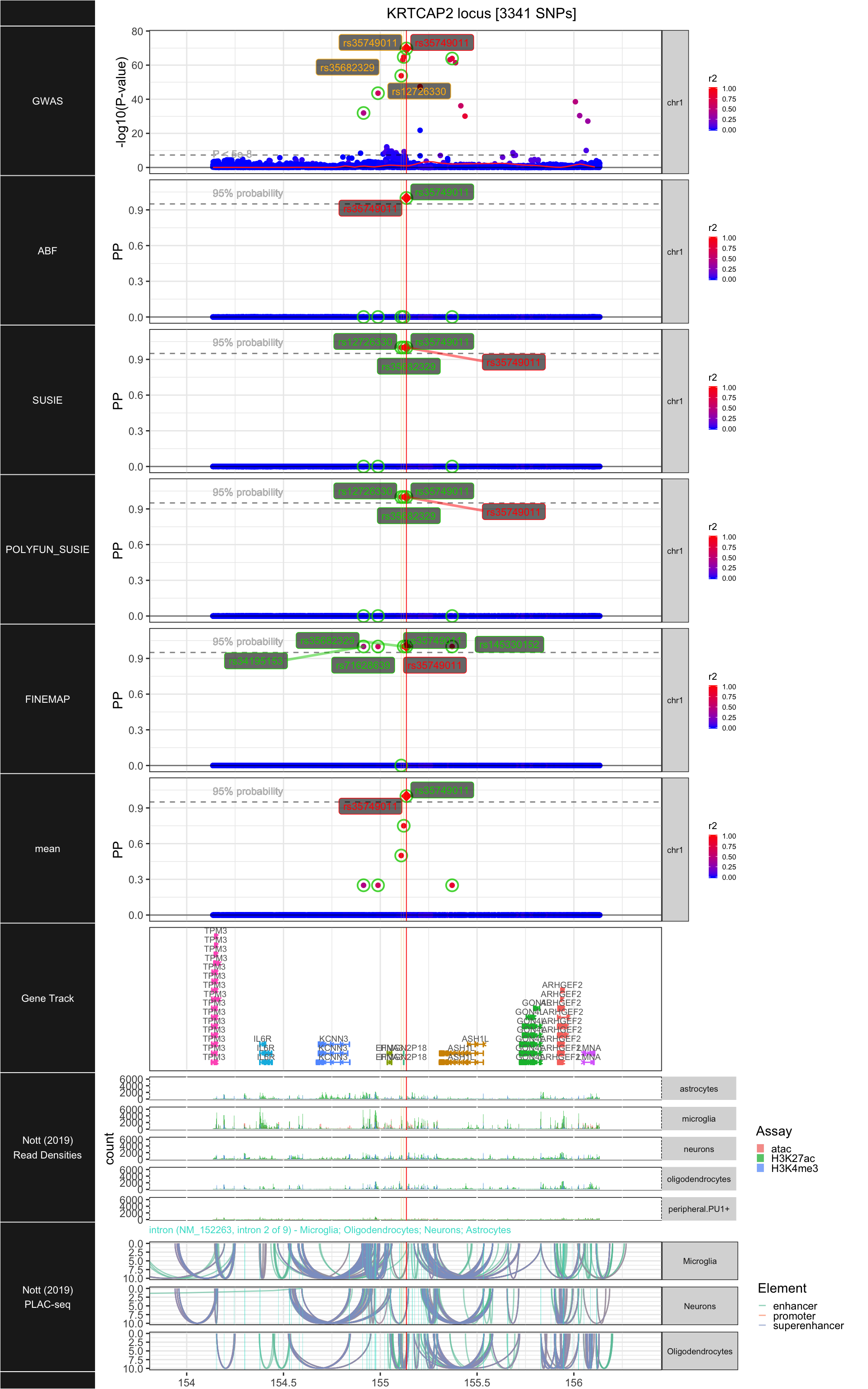
## ### KRTCAP2
## 
##
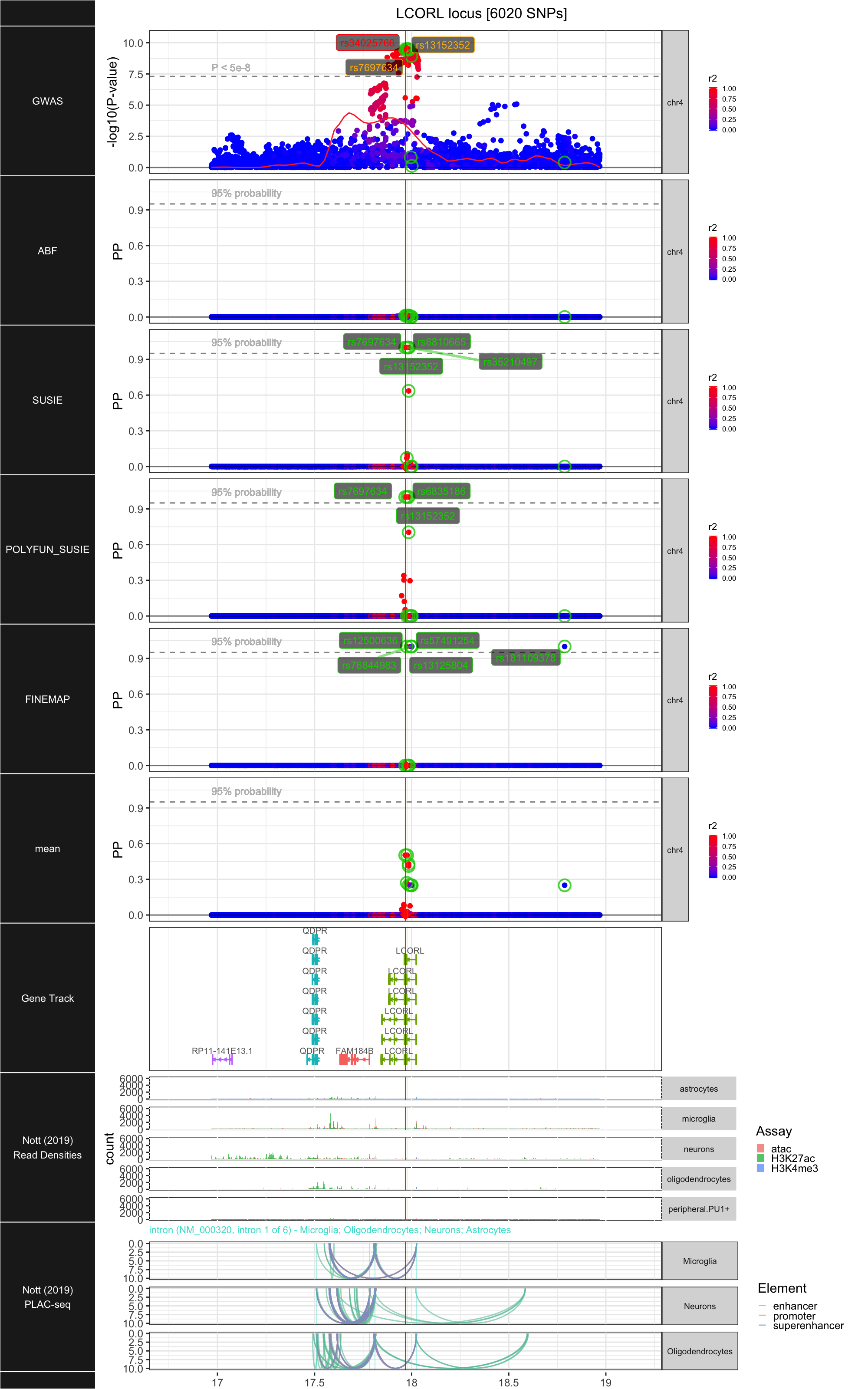
## ### LCORL
## 
##
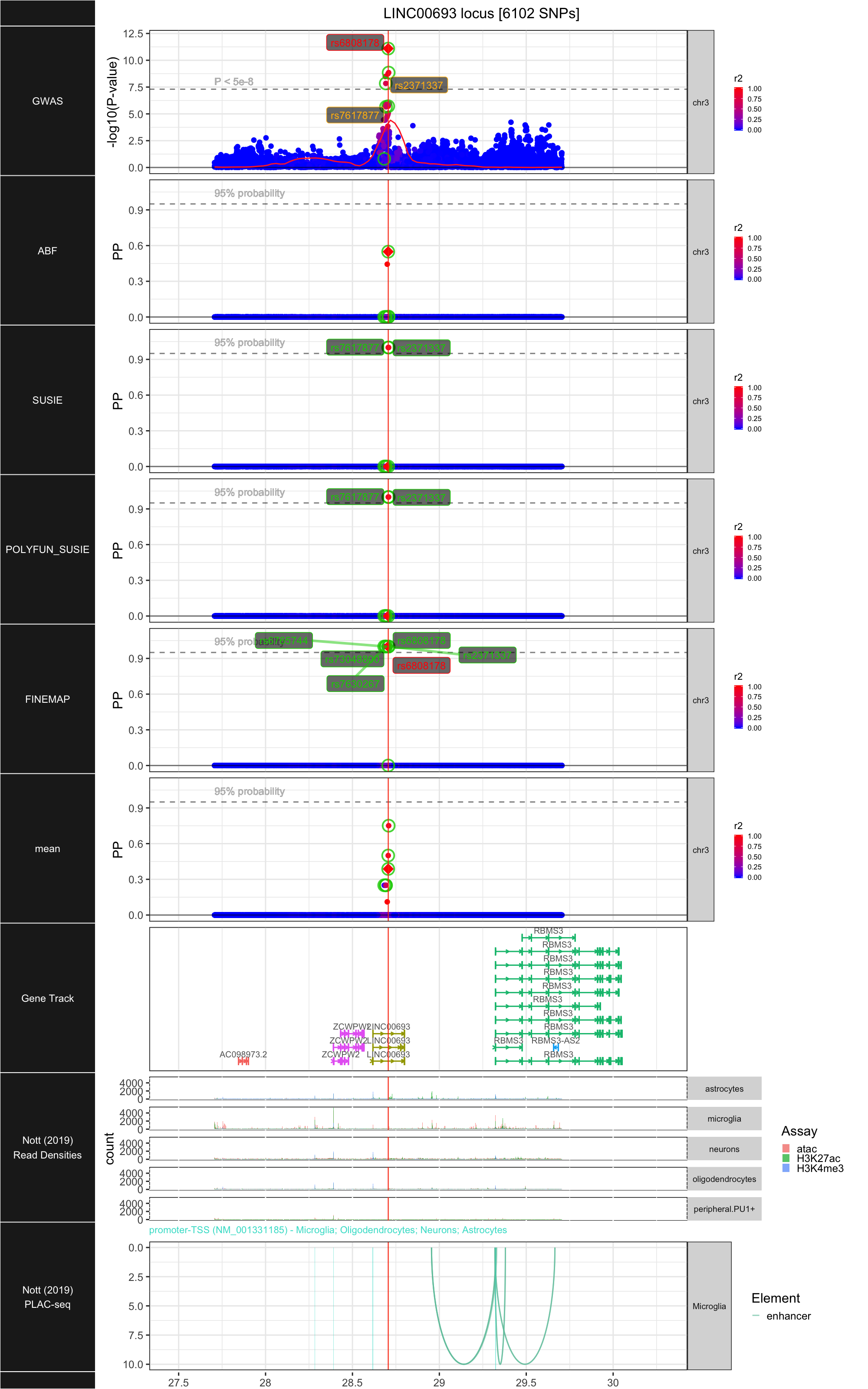
## ### LINC00693
## 
##
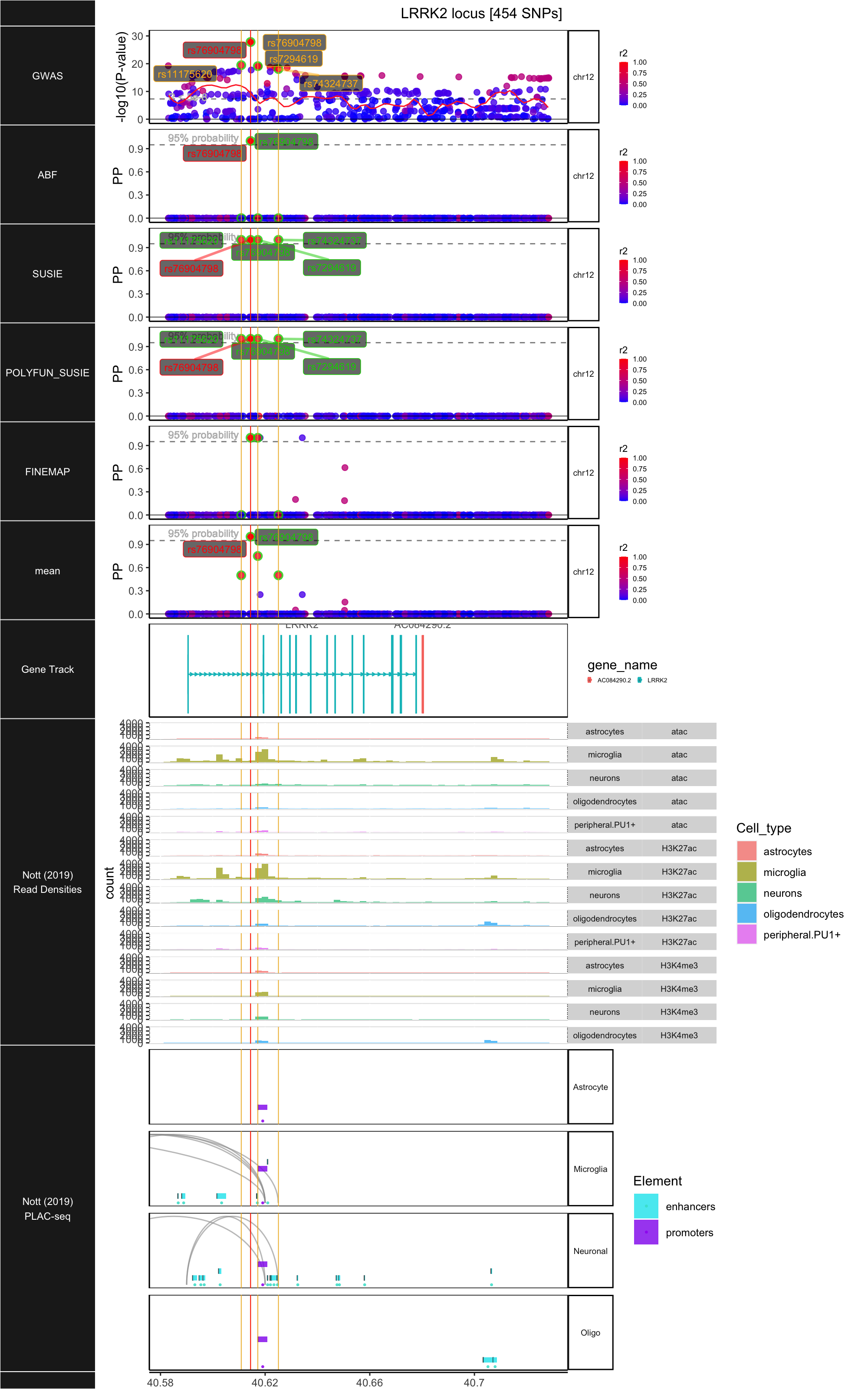
## ### LRRK2
## 
##
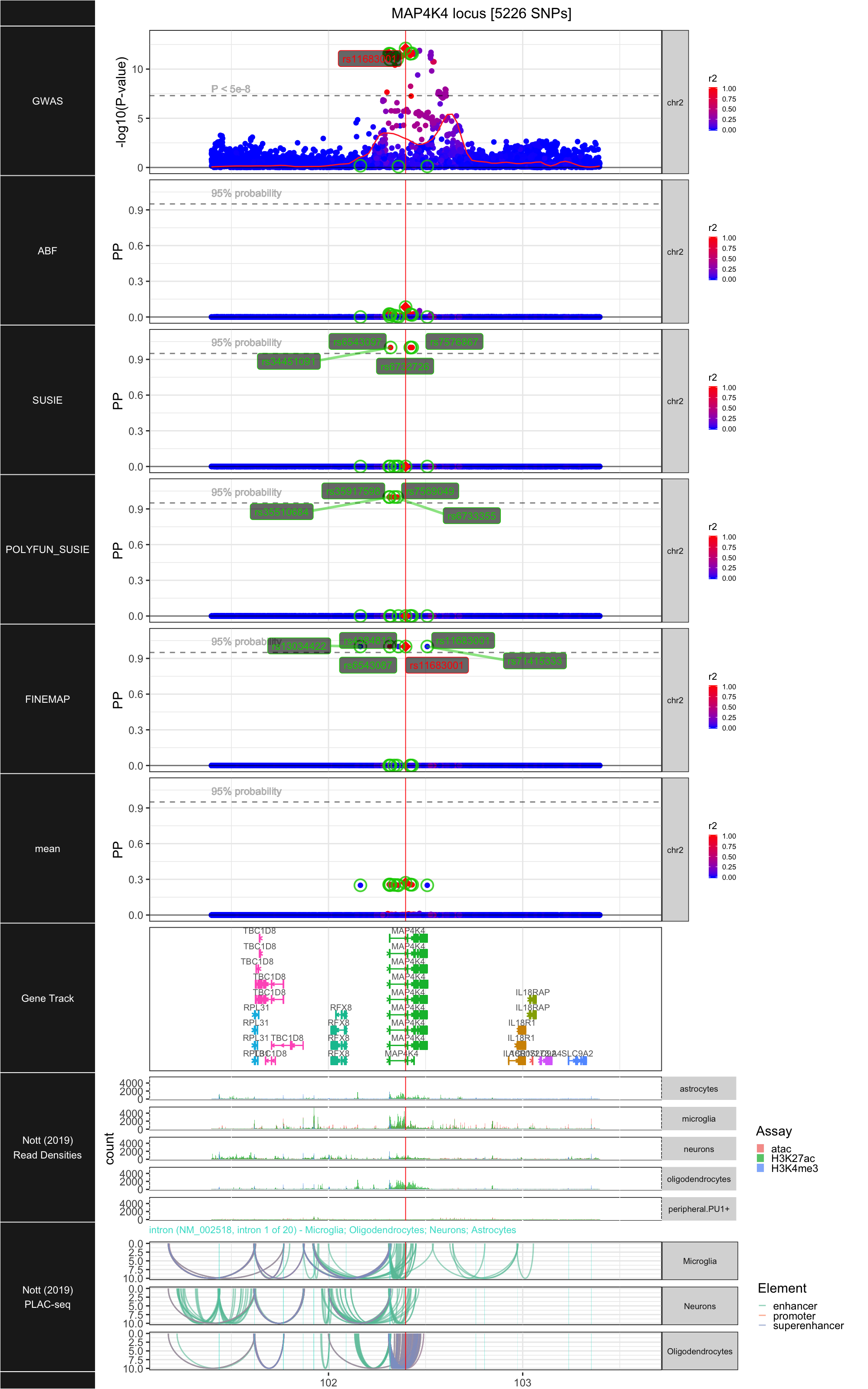
## ### MAP4K4
## 
##
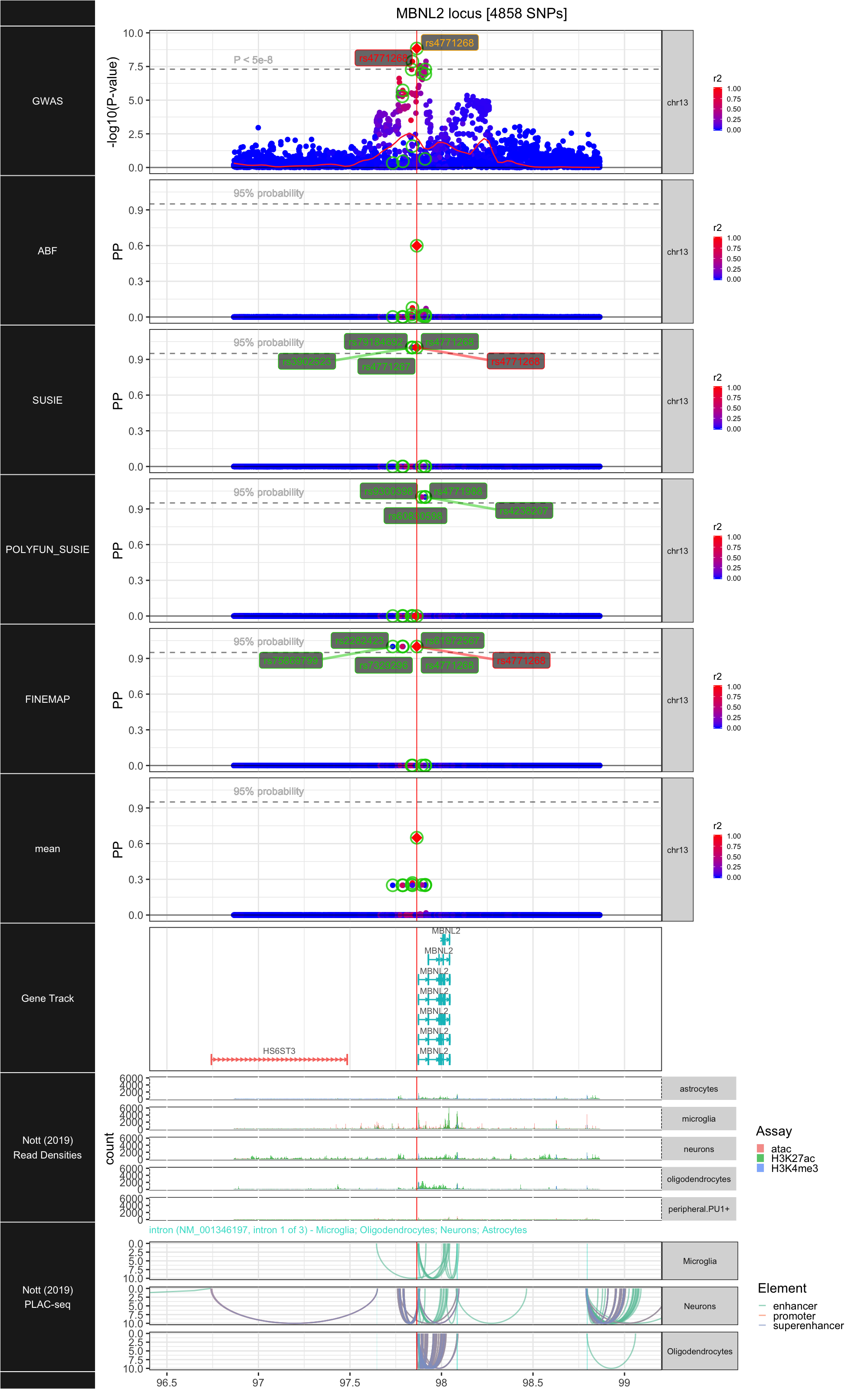
## ### MBNL2
## 
##
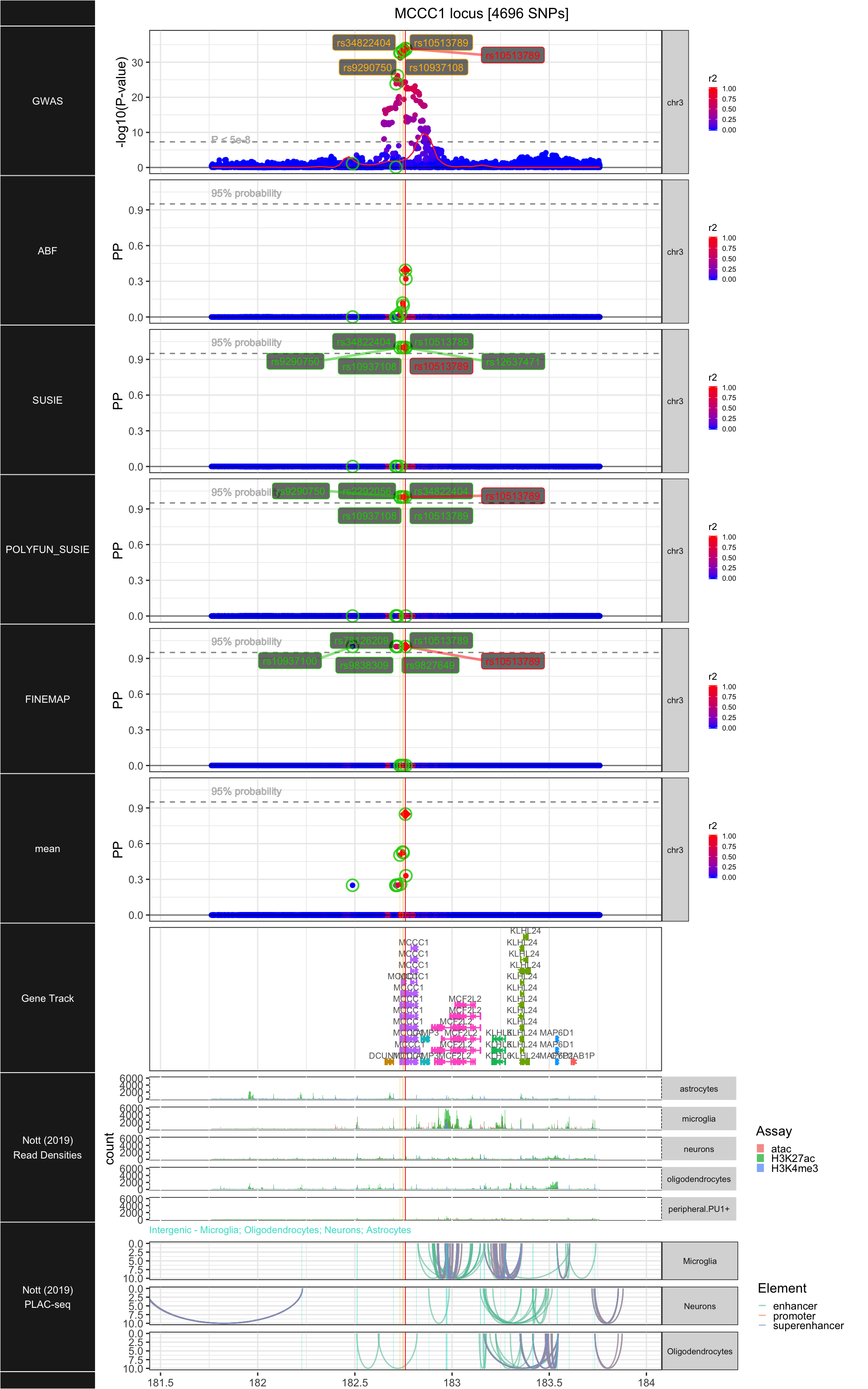
## ### MCCC1
## 
##
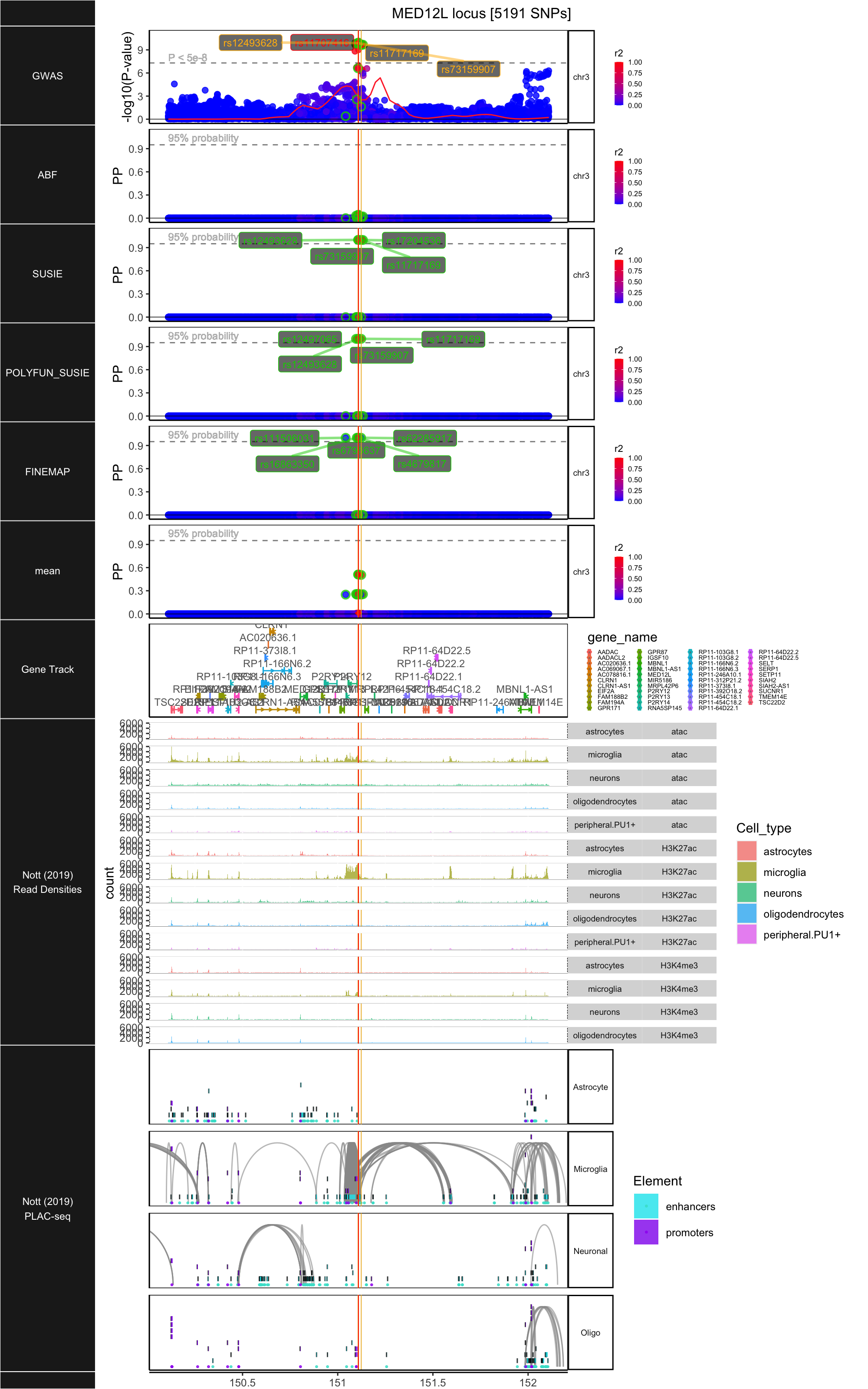
## ### MED12L
## 
##
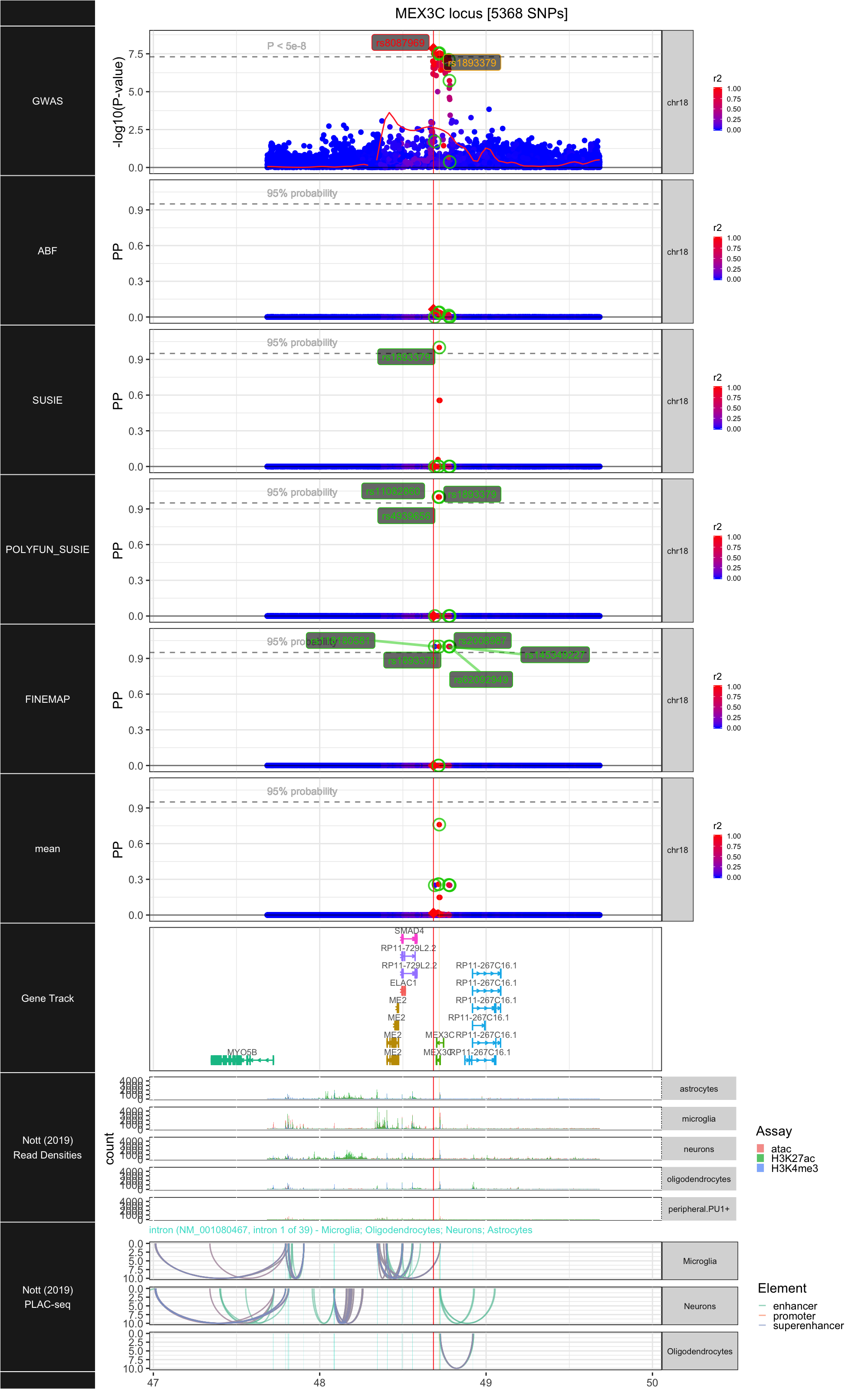
## ### MEX3C
## 
##
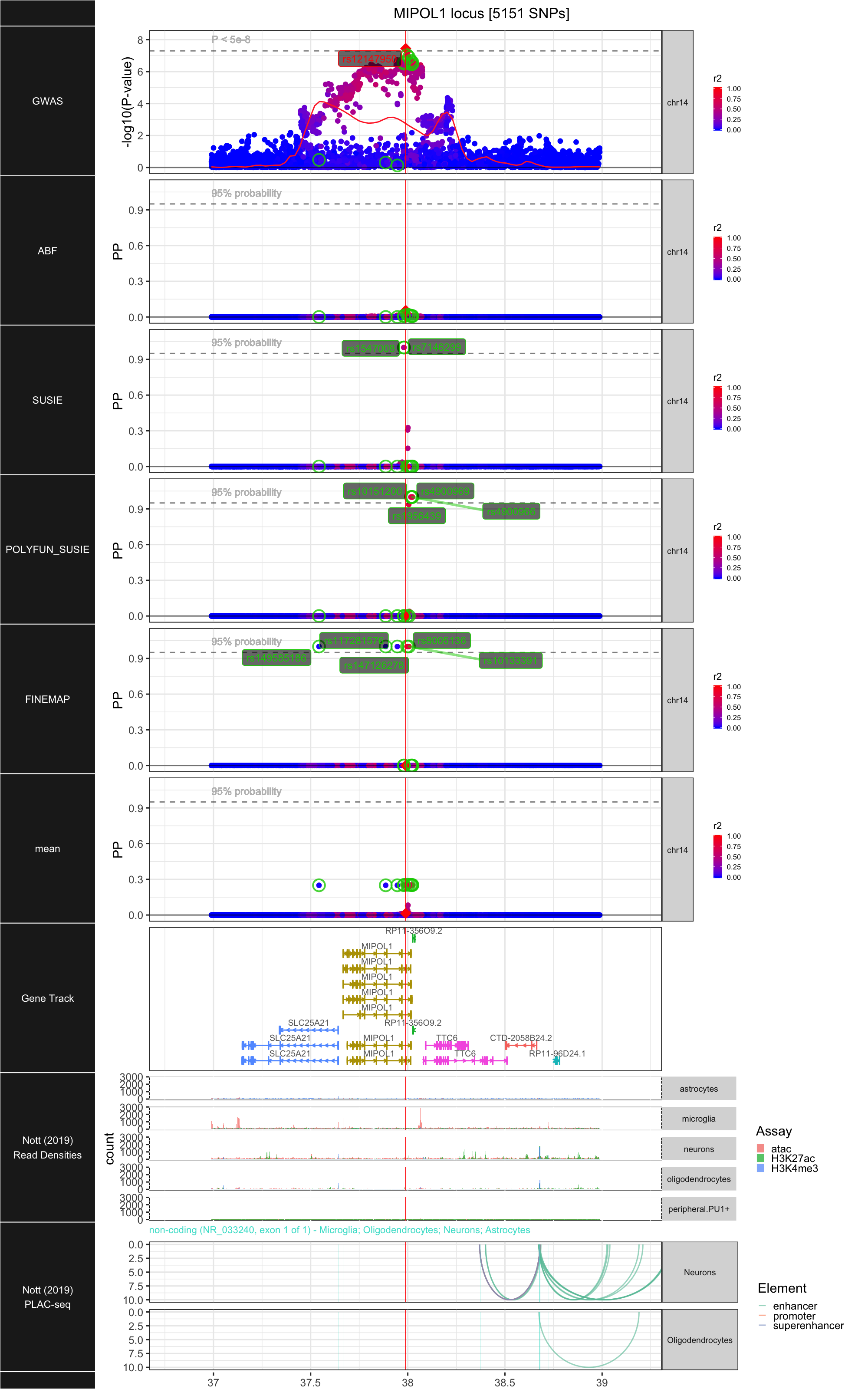
## ### MIPOL1
## 
##
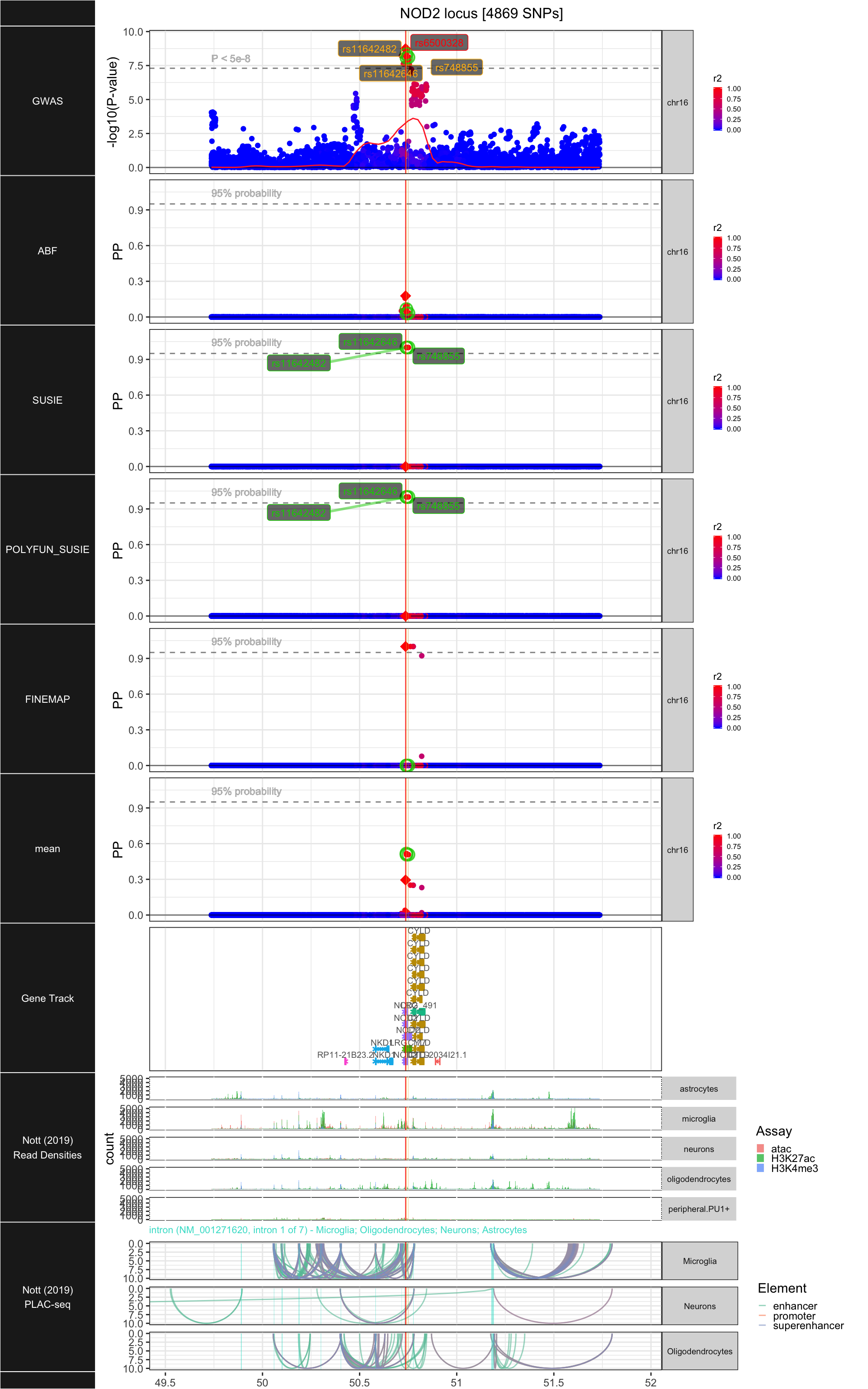
## ### NOD2
## 
##
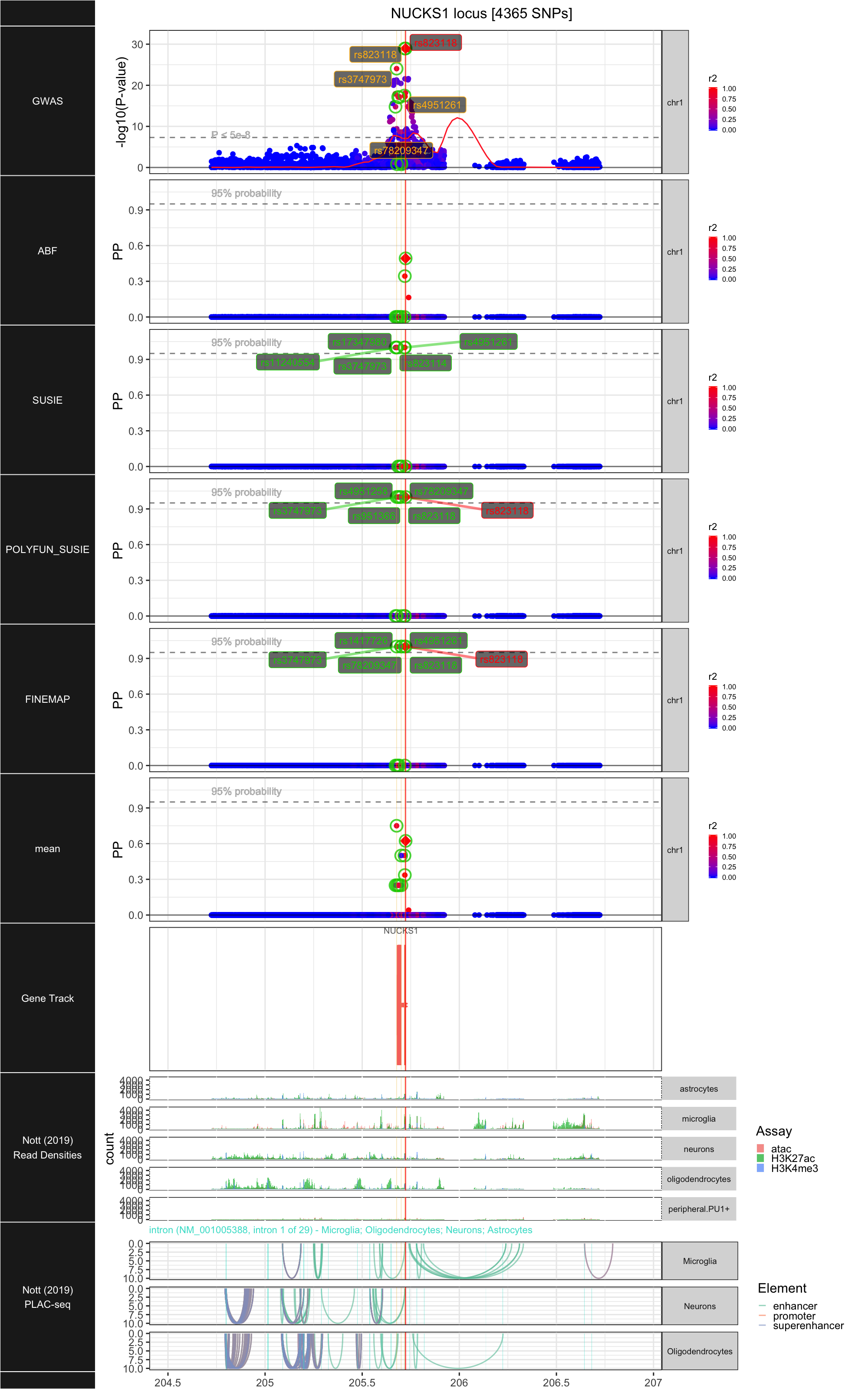
## ### NUCKS1
## 
##
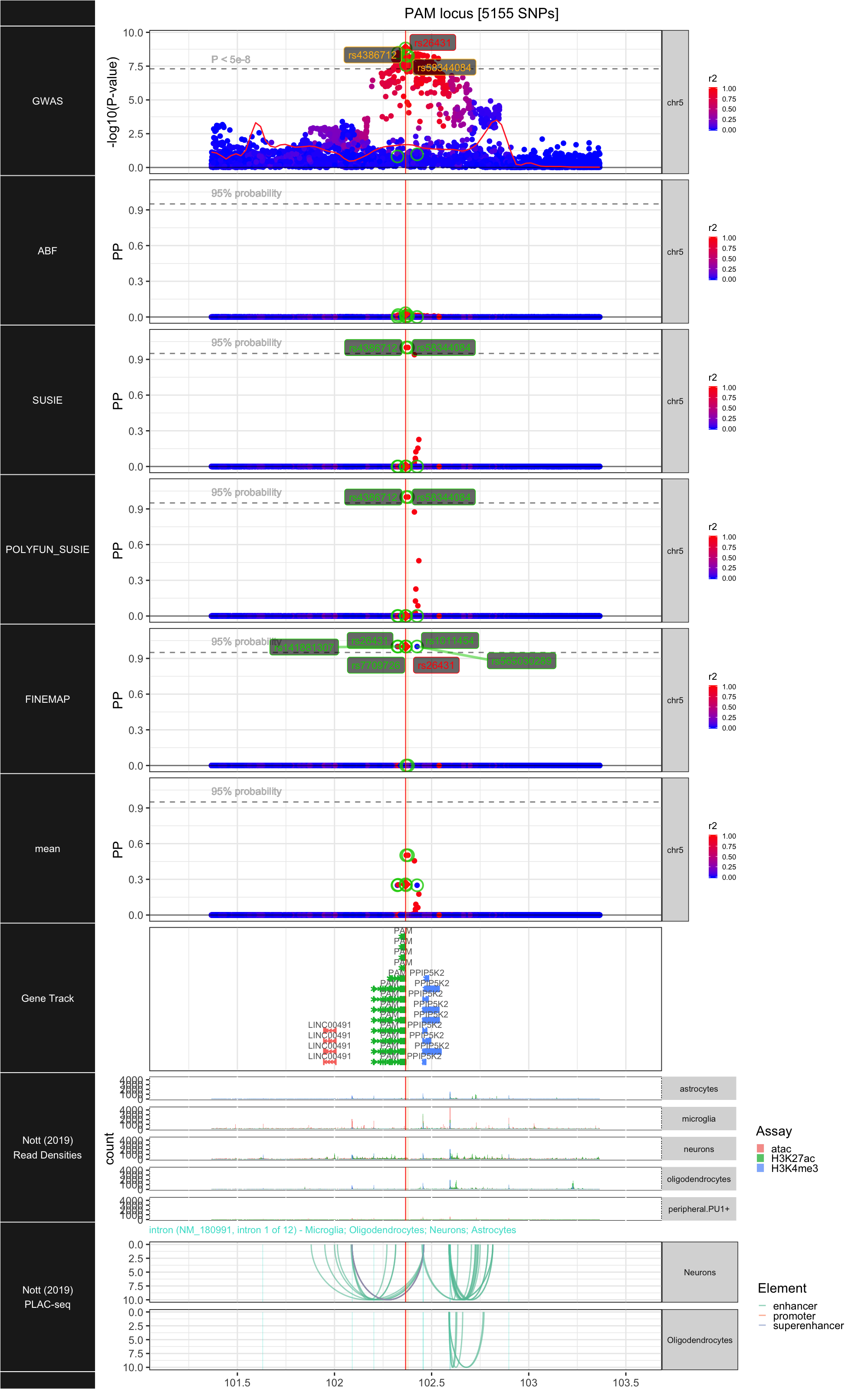
## ### PAM
## 
##
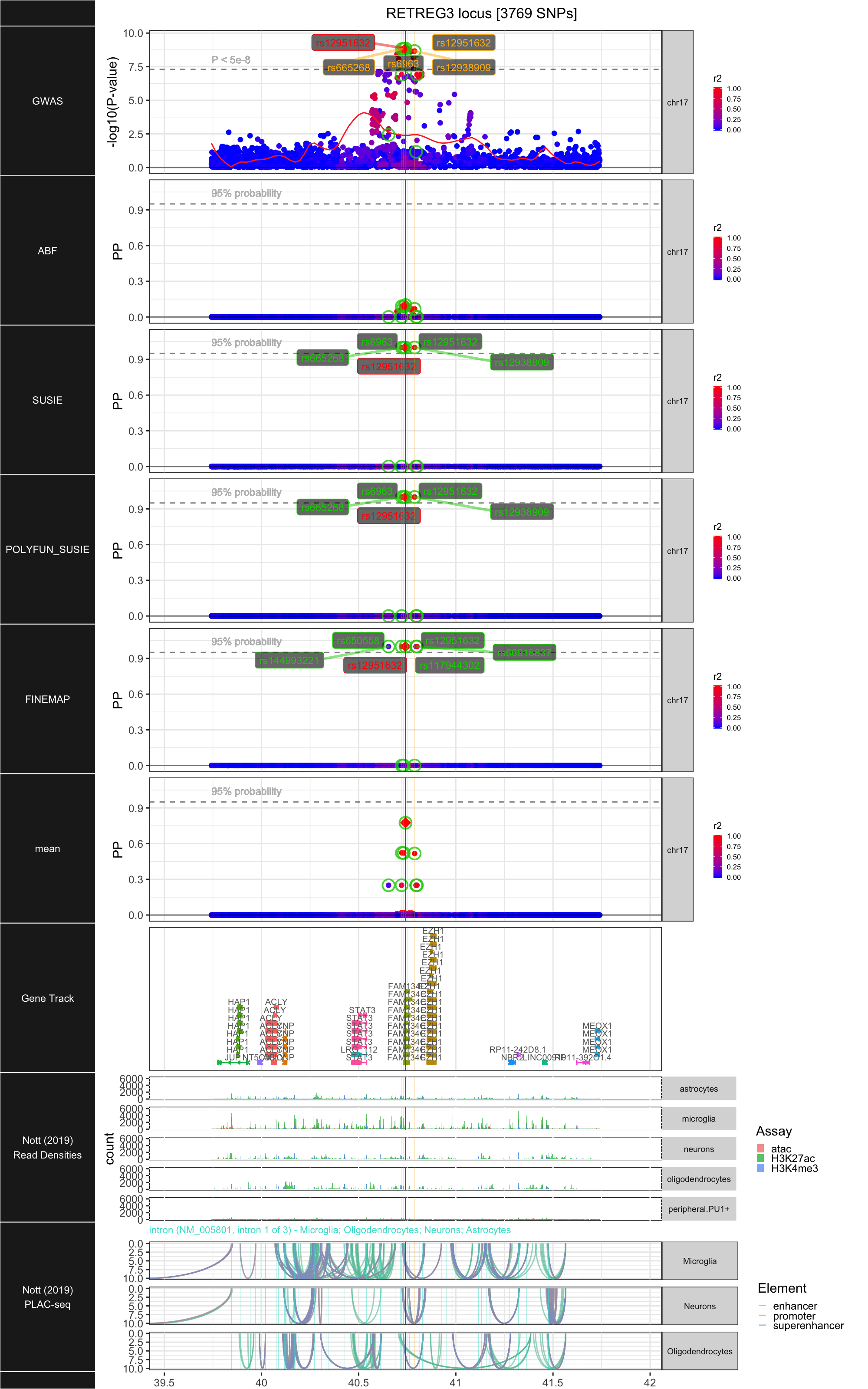
## ### RETREG3
## 
##
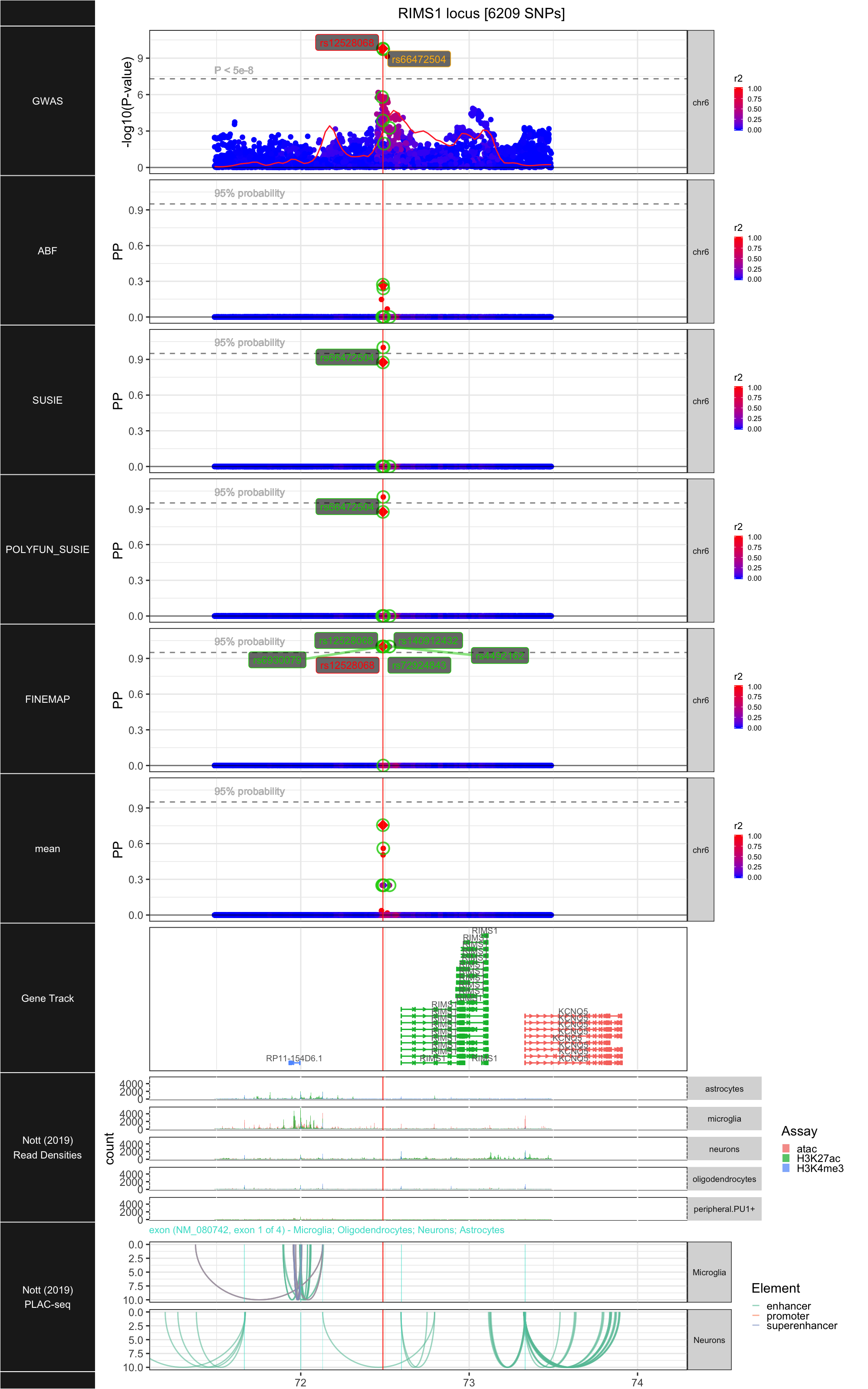
## ### RIMS1
## 
##
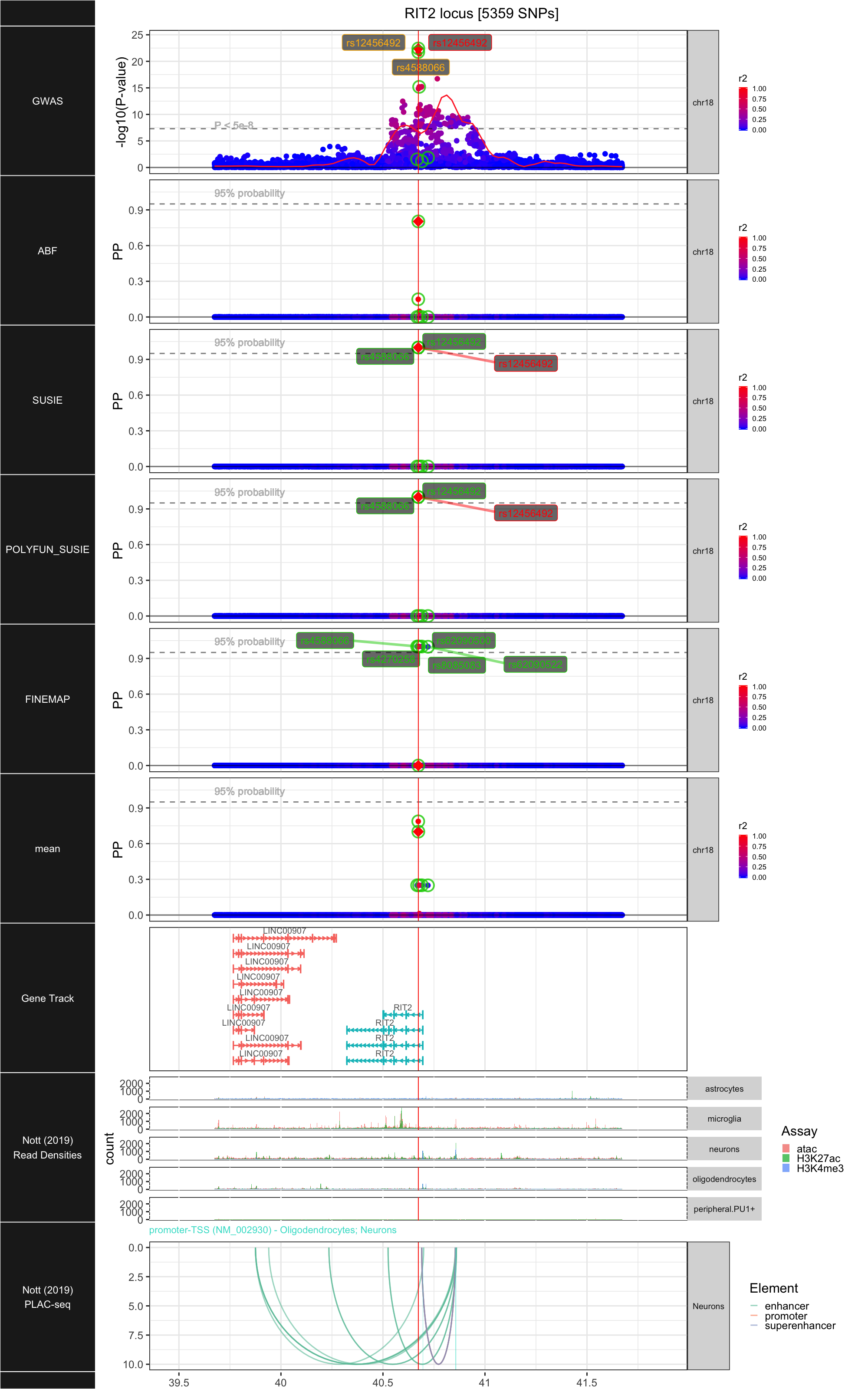
## ### RIT2
## 
##
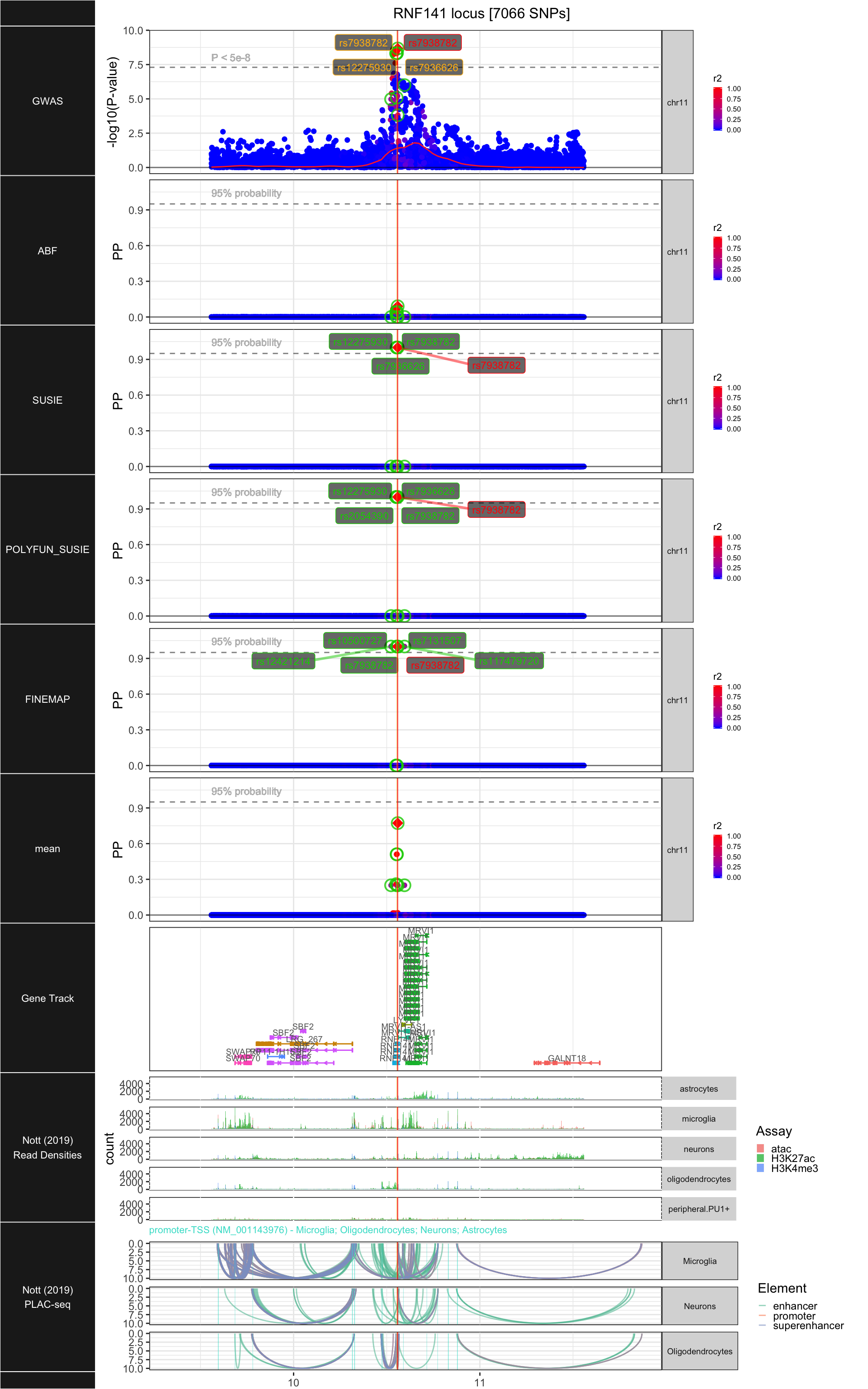
## ### RNF141
## 
##
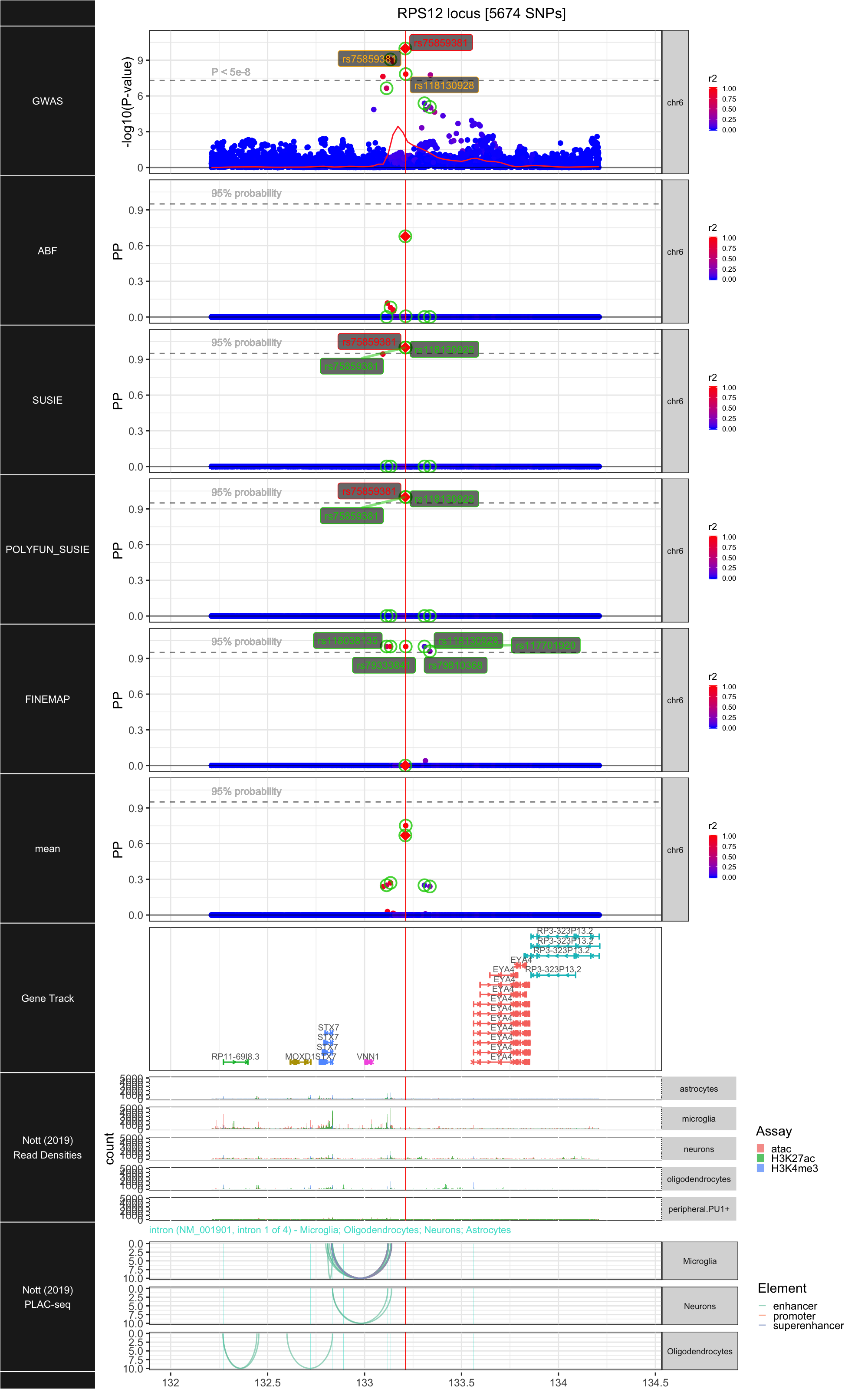
## ### RPS12
## 
##
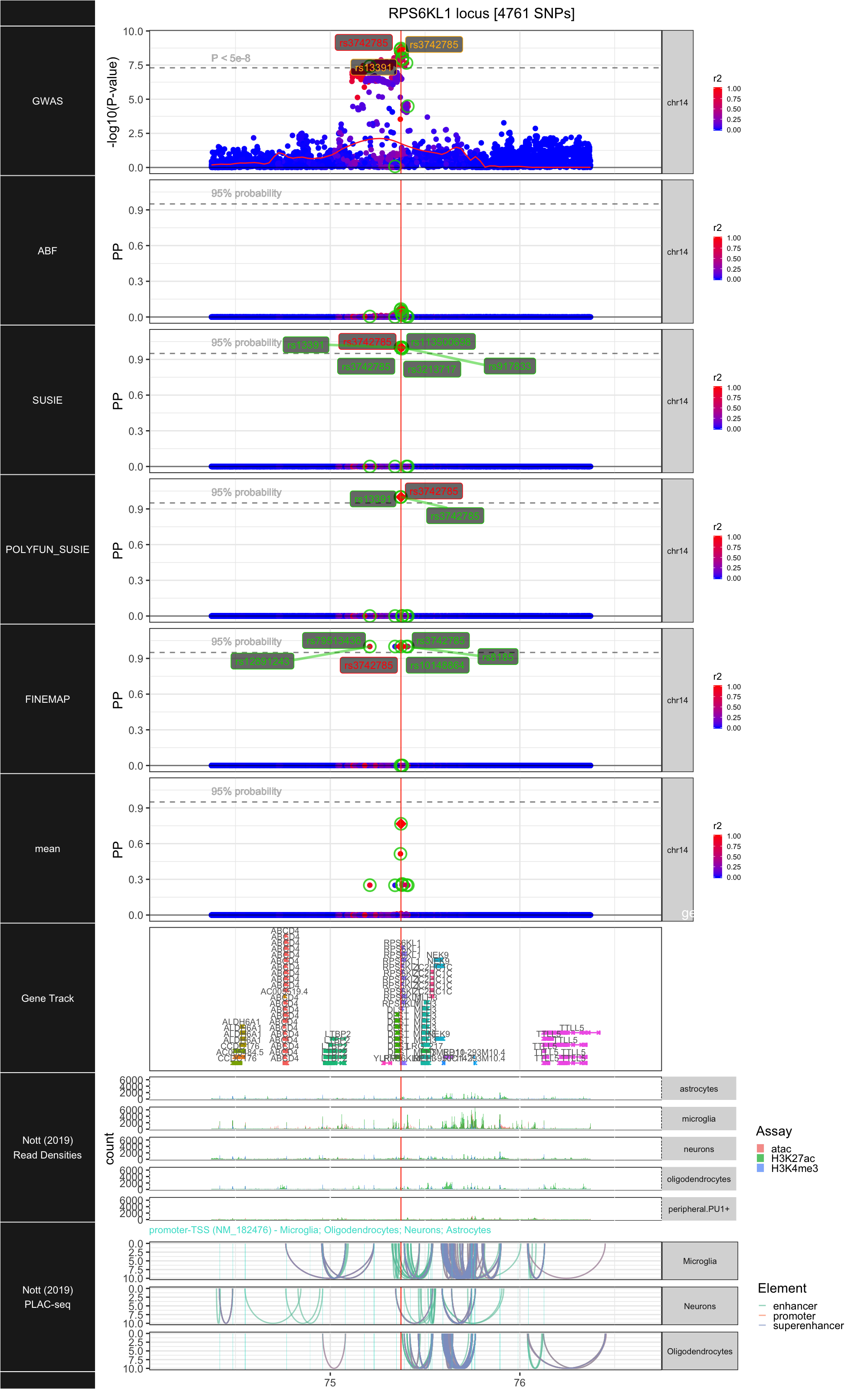
## ### RPS6KL1
## 
##
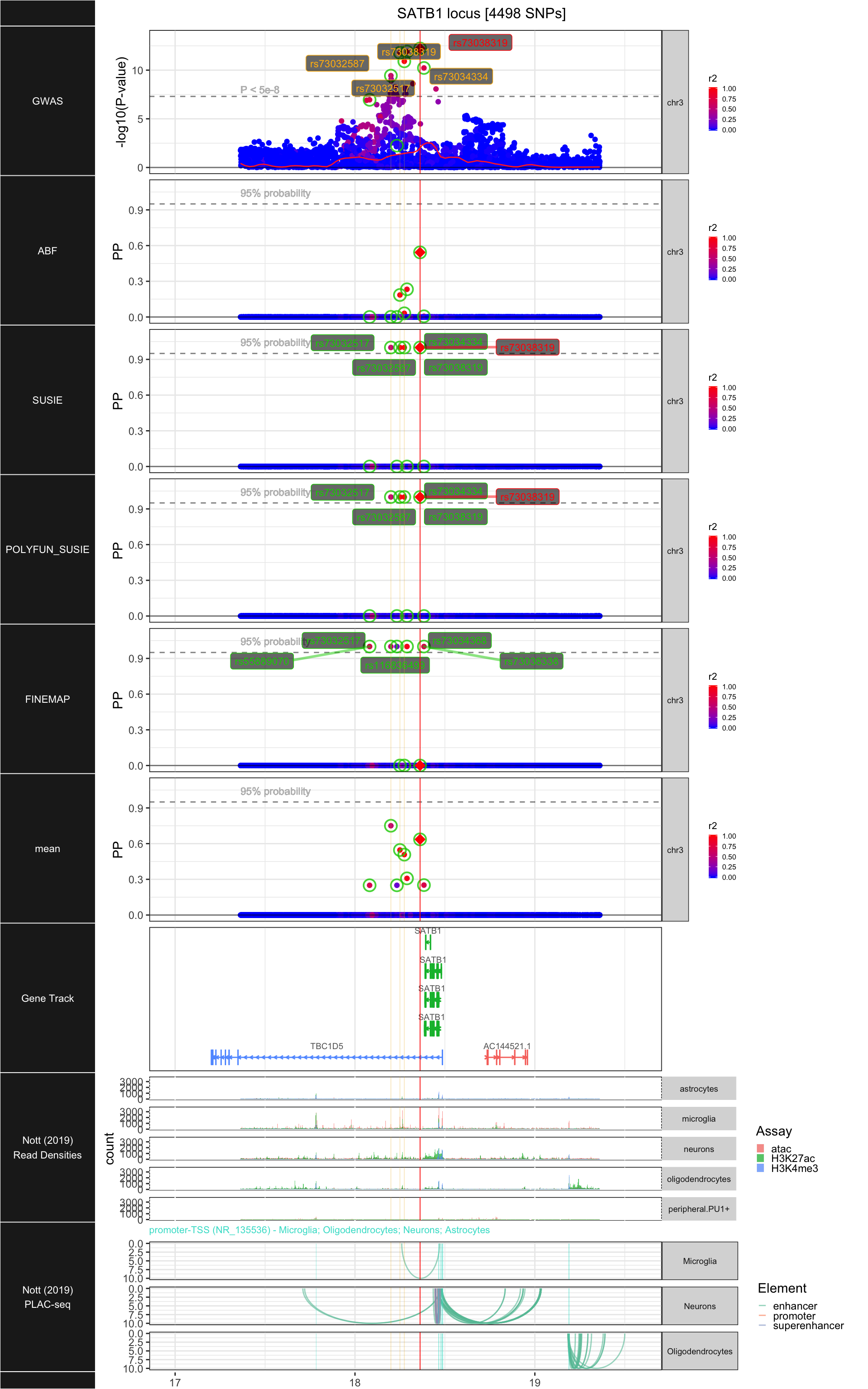
## ### SATB1
## 
##
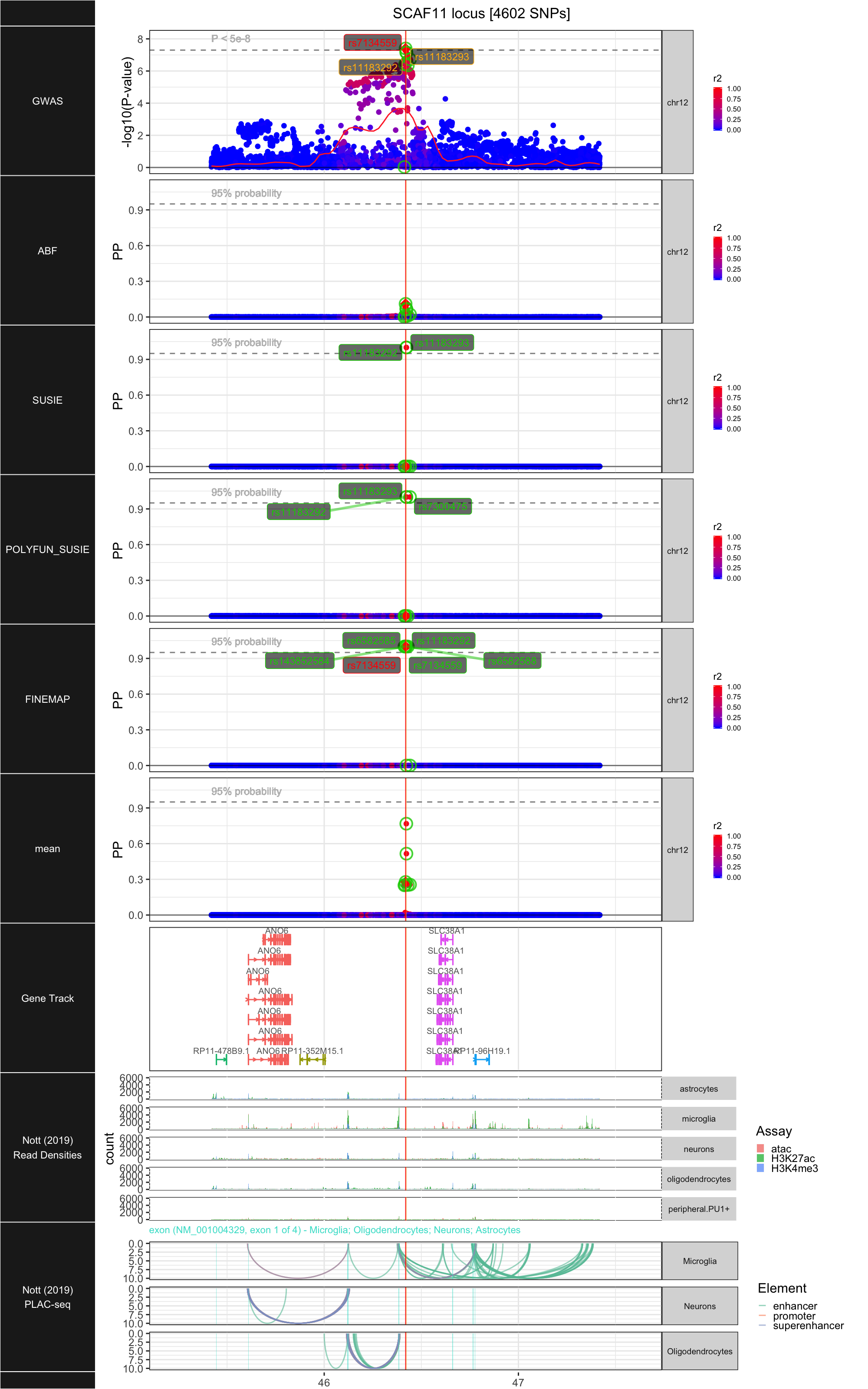
## ### SCAF11
## 
##
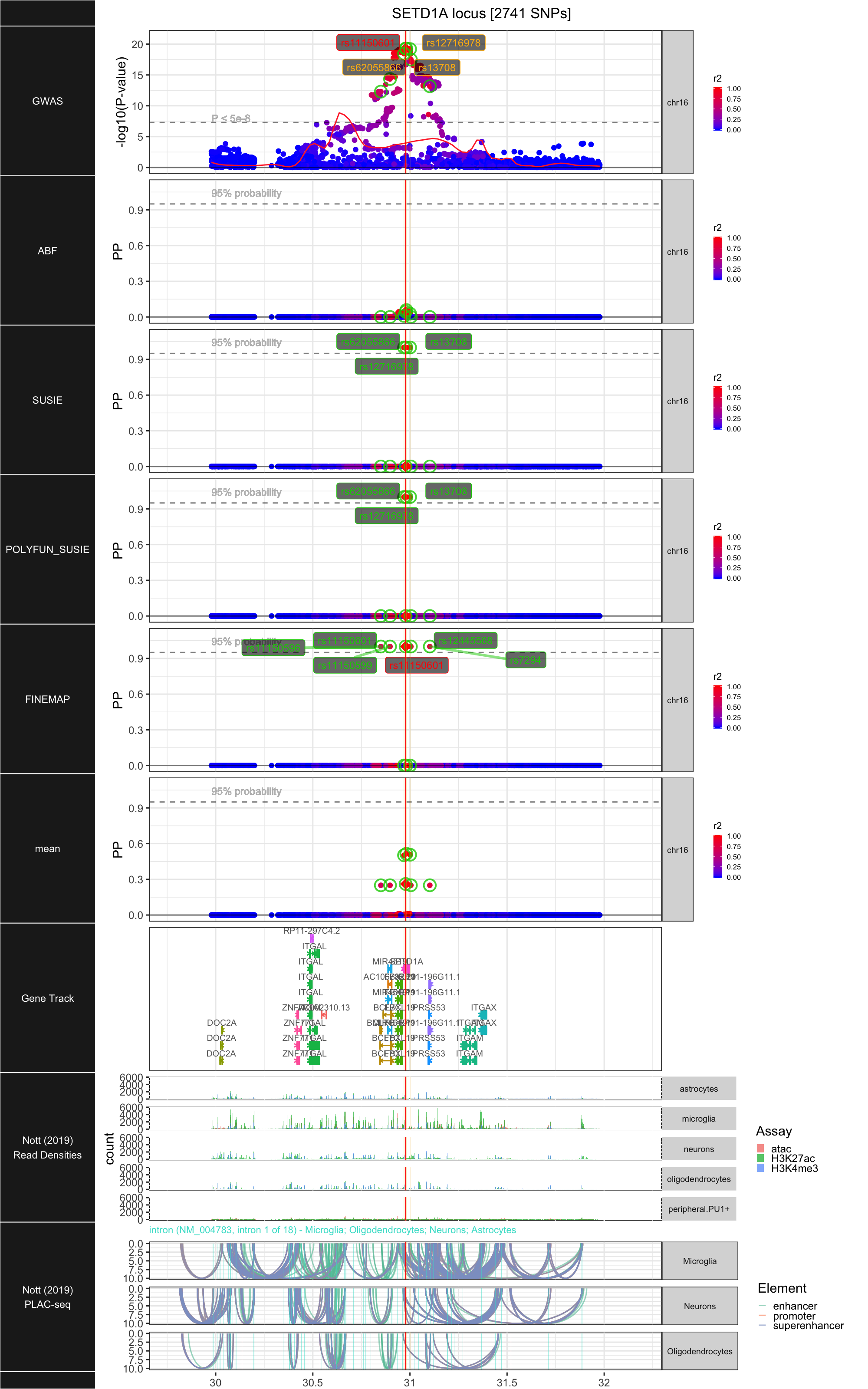
## ### SETD1A
## 
##
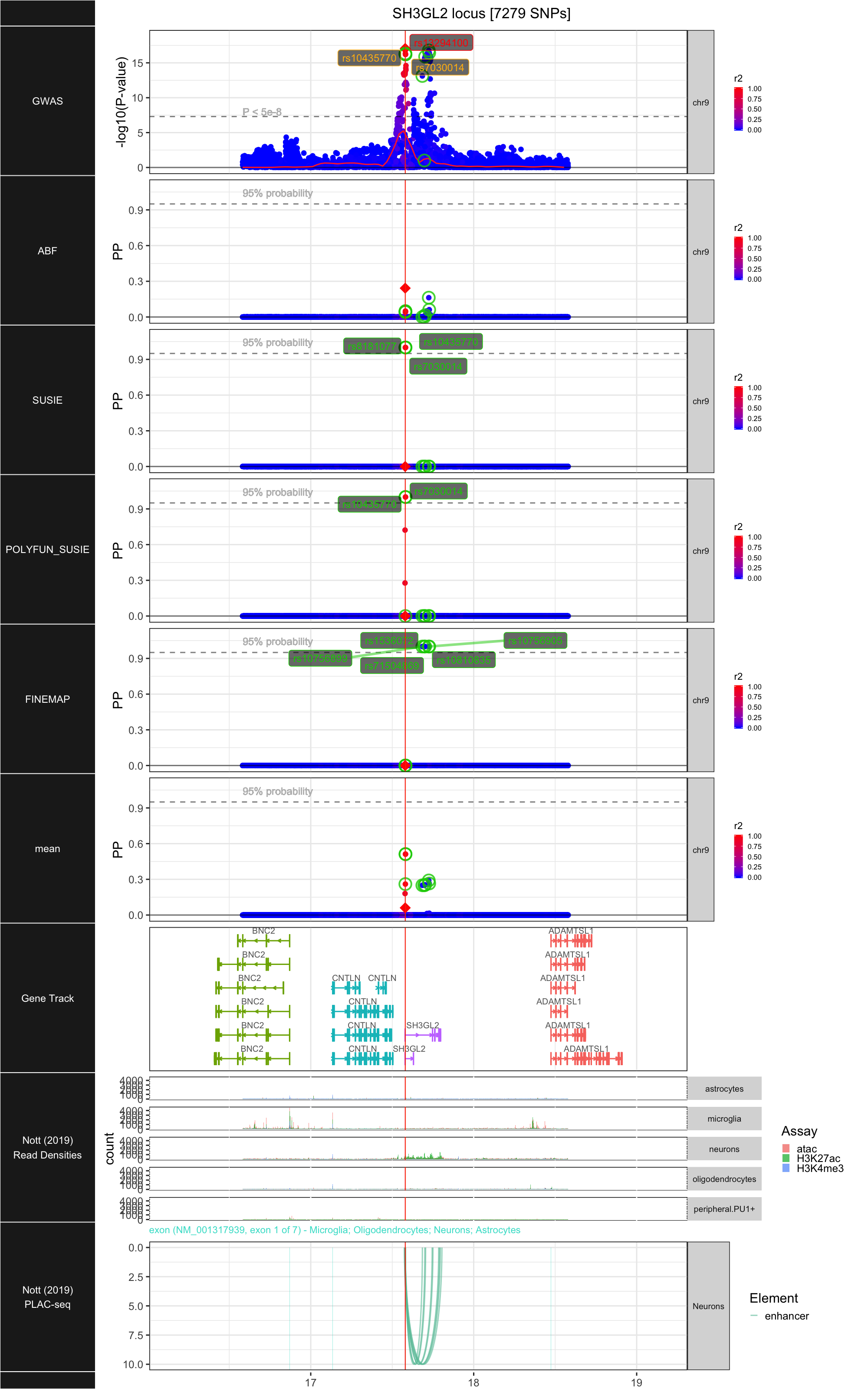
## ### SH3GL2
## 
##
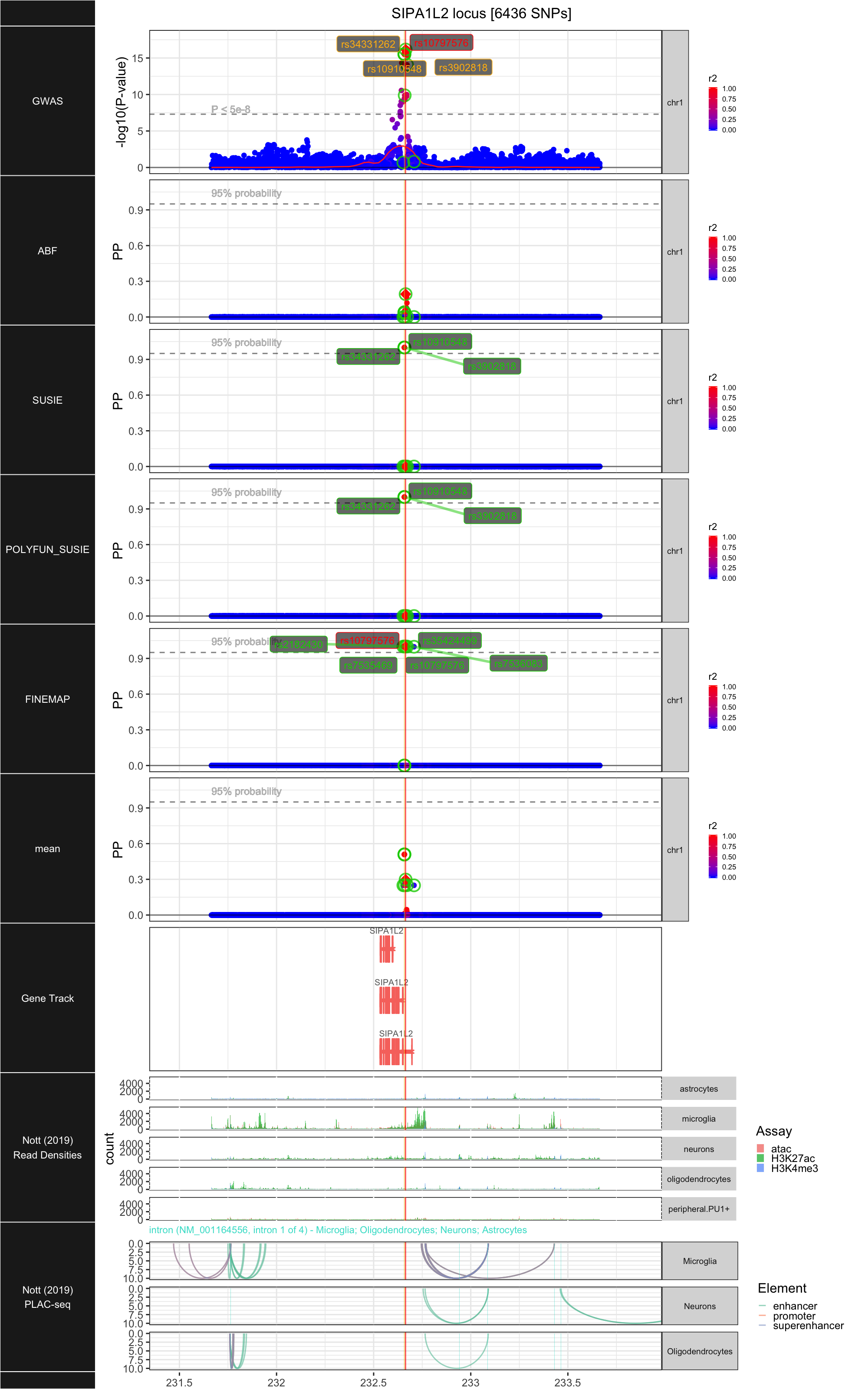
## ### SIPA1L2
## 
##
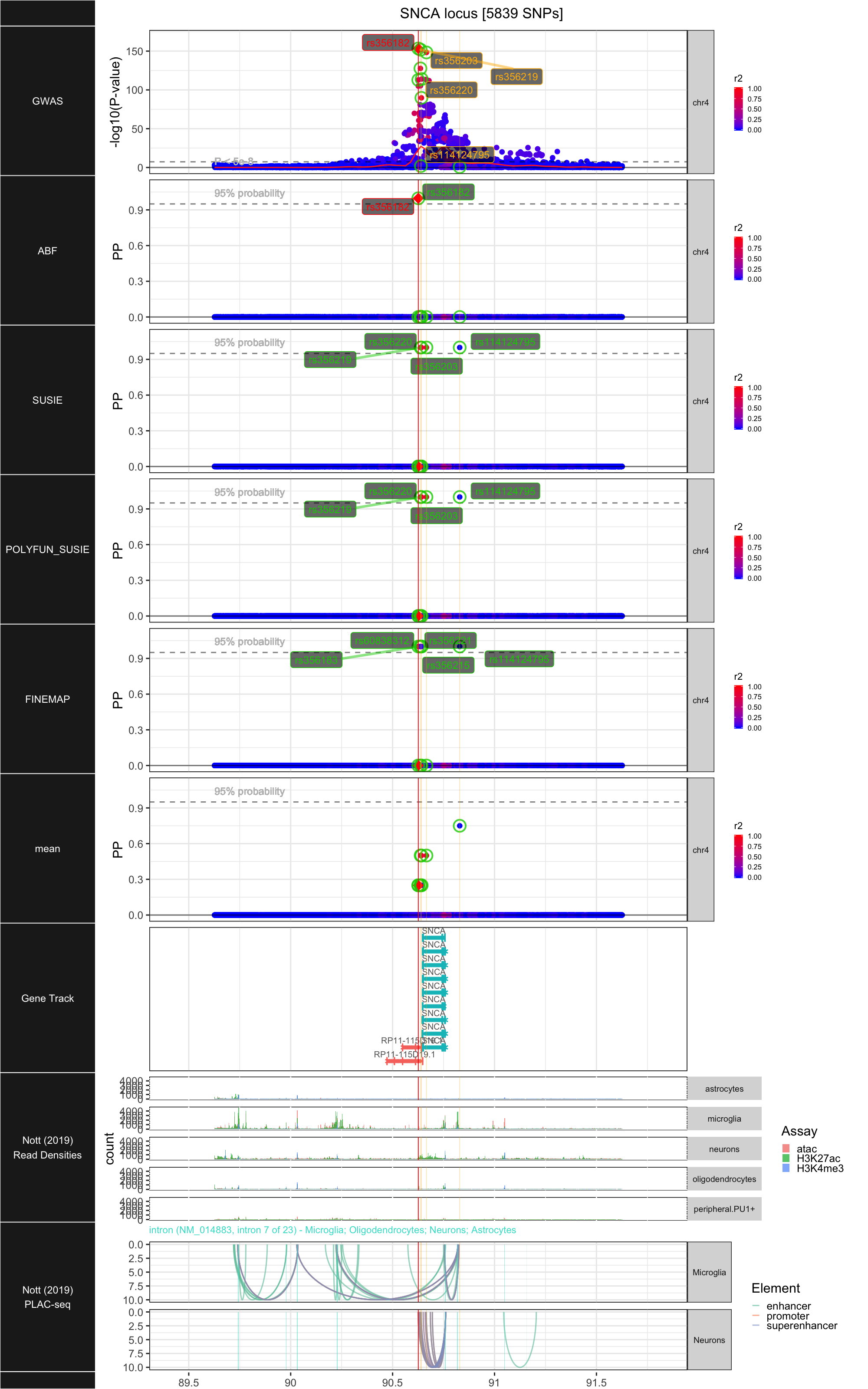
## ### SNCA
## 
##
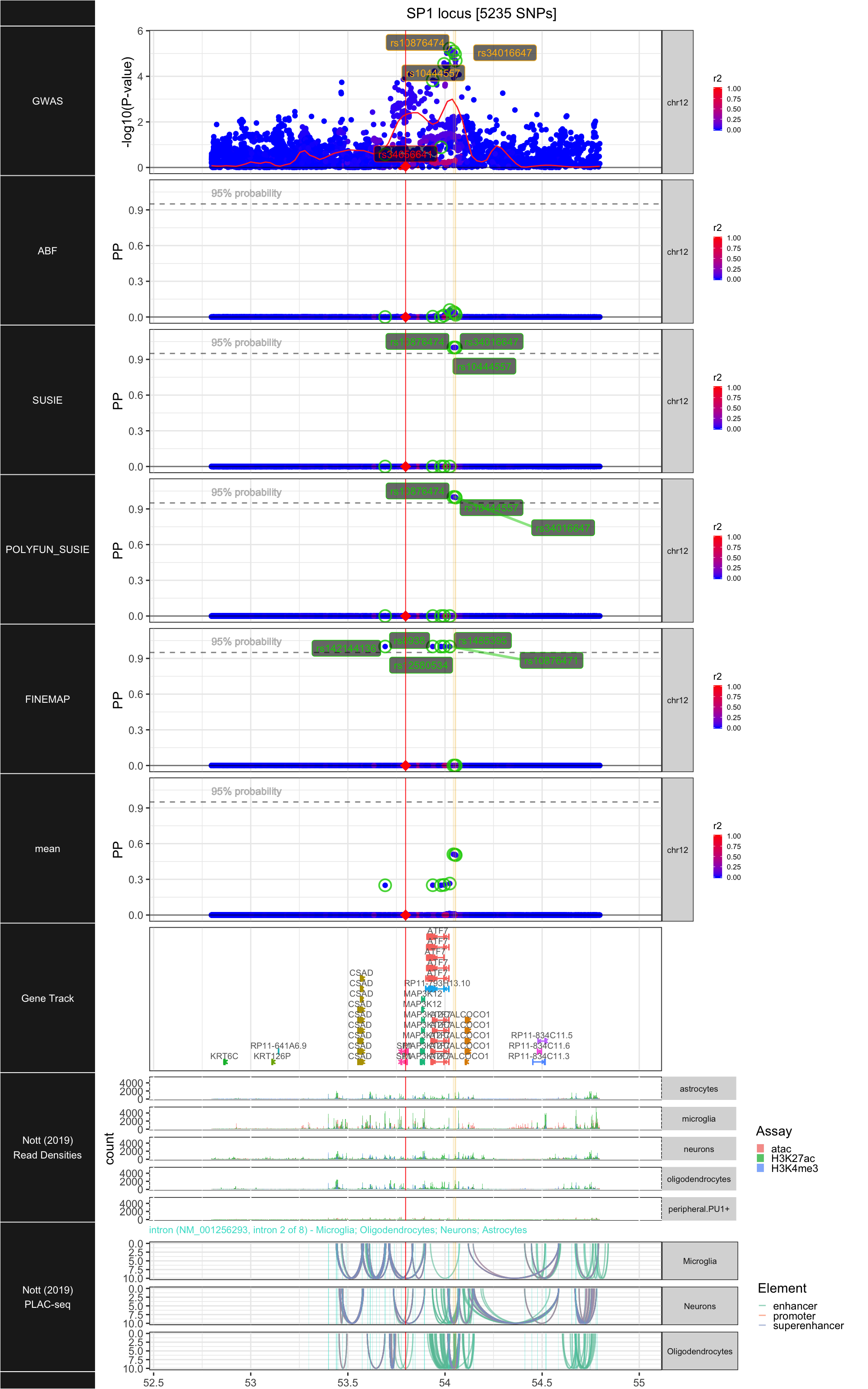
## ### SP1
## 
##
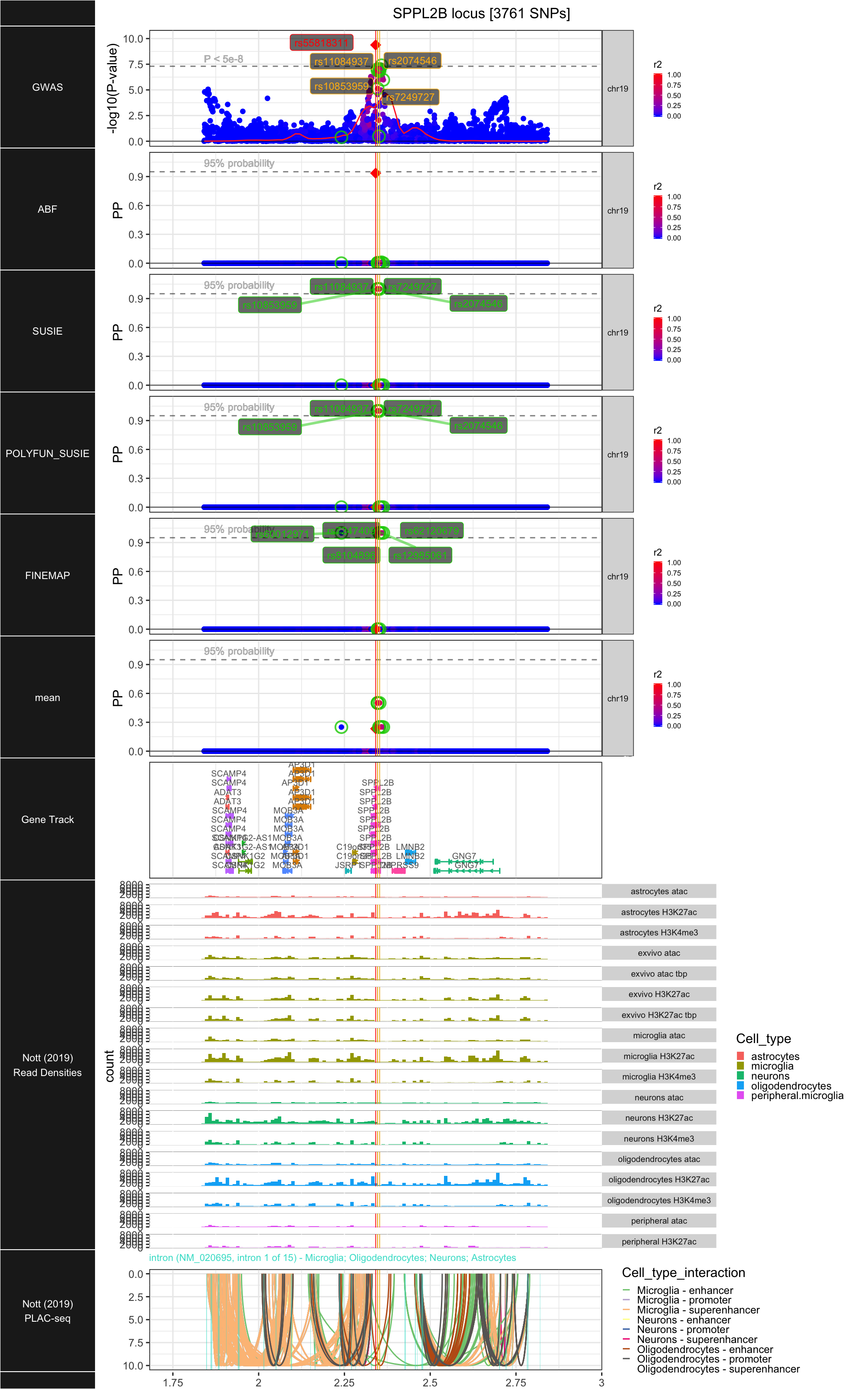
## ### SPPL2B
## 
##
## ### SPTSSB
## 
##
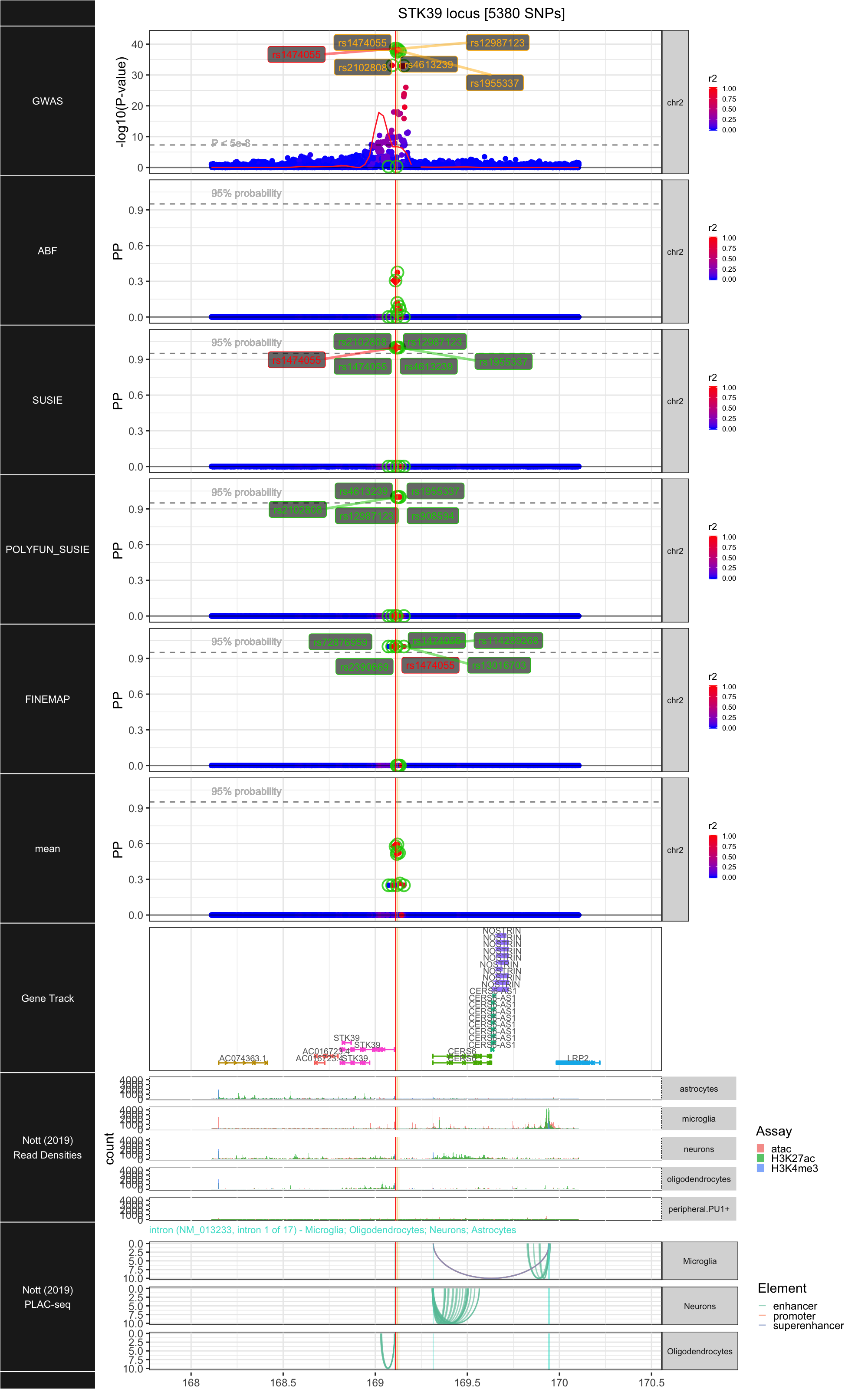
## ### STK39
## 
##
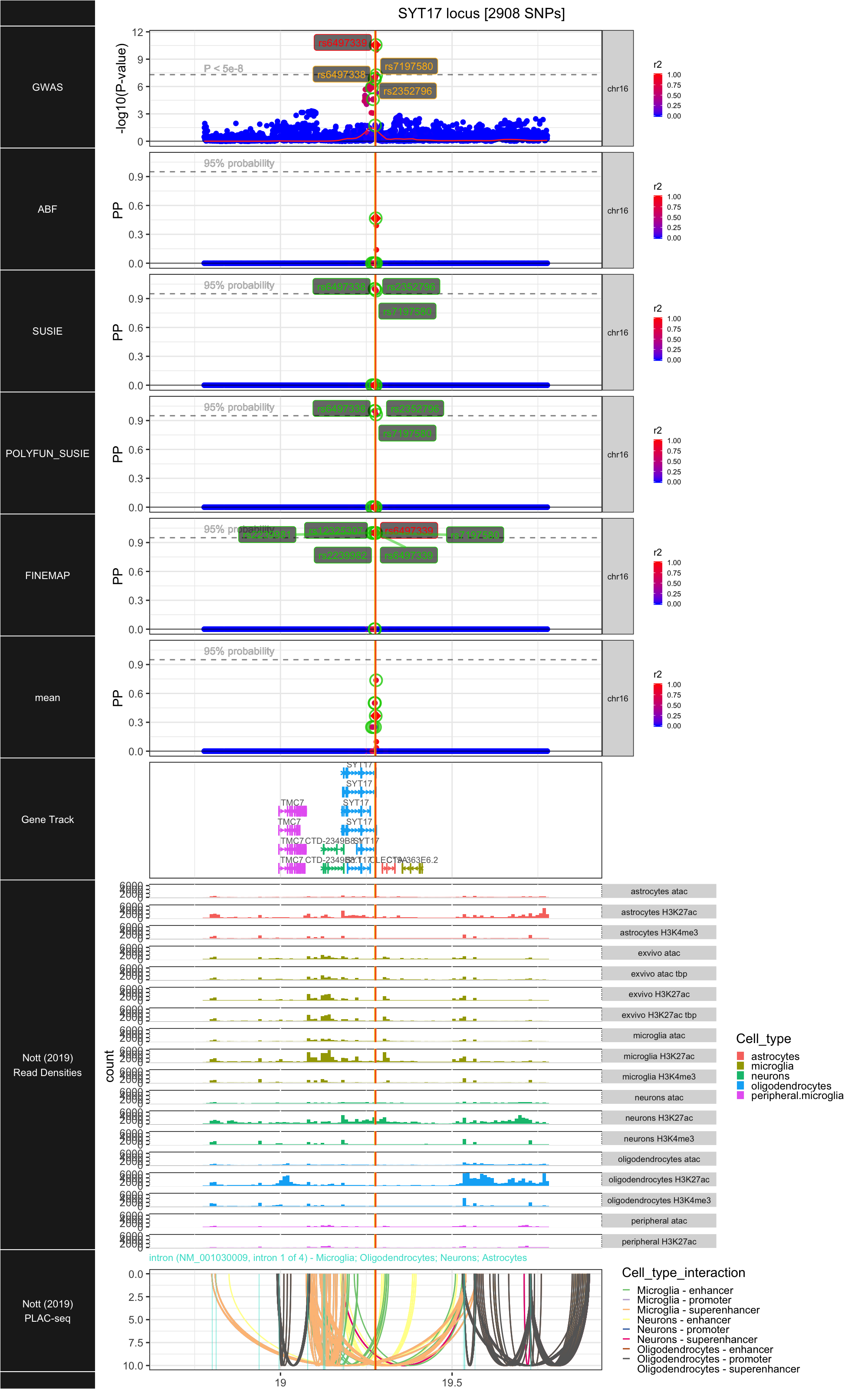
## ### SYT17
## 
##
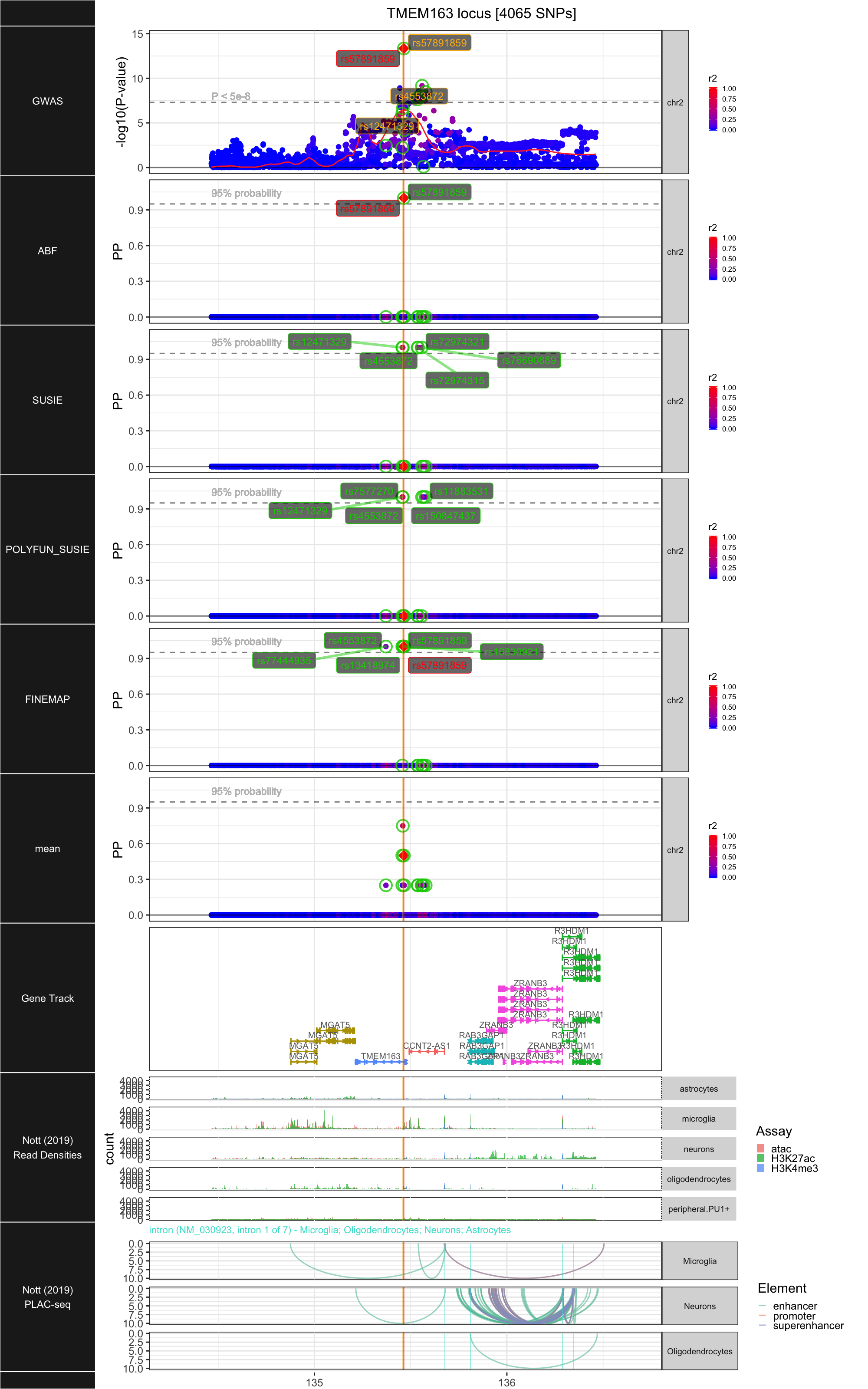
## ### TMEM163
## 
##
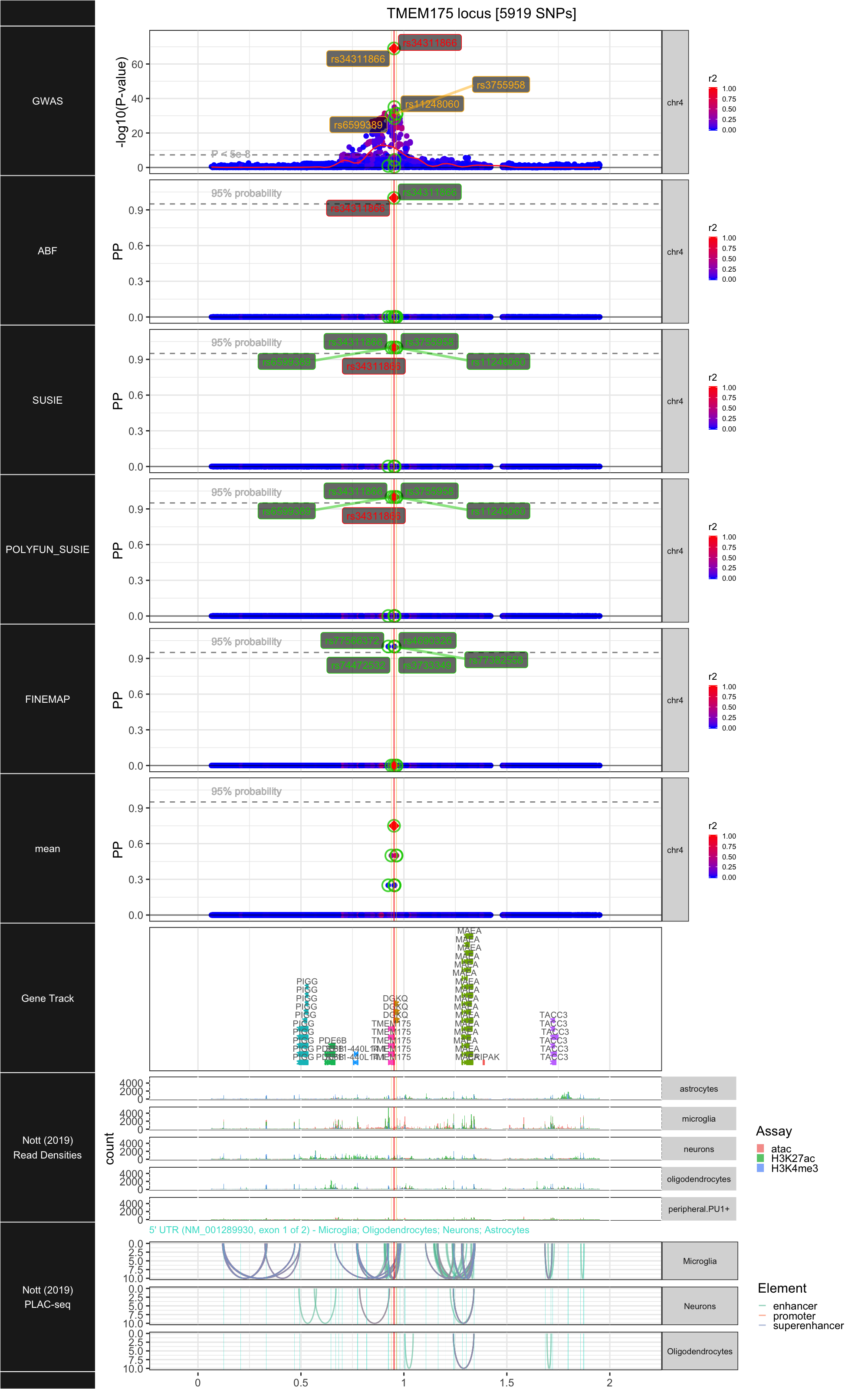
## ### TMEM175
## 
##
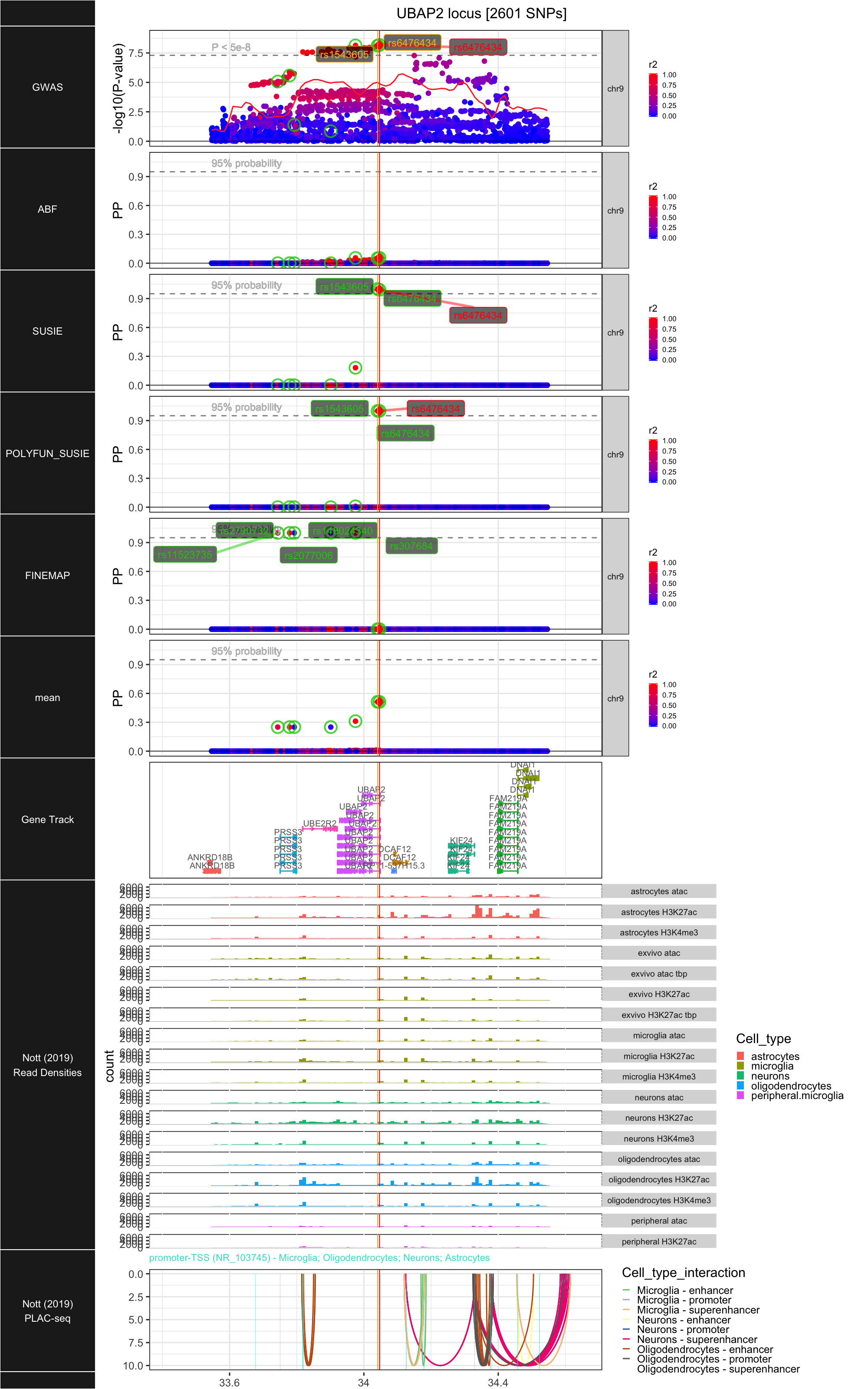
## ### UBAP2
## 
##
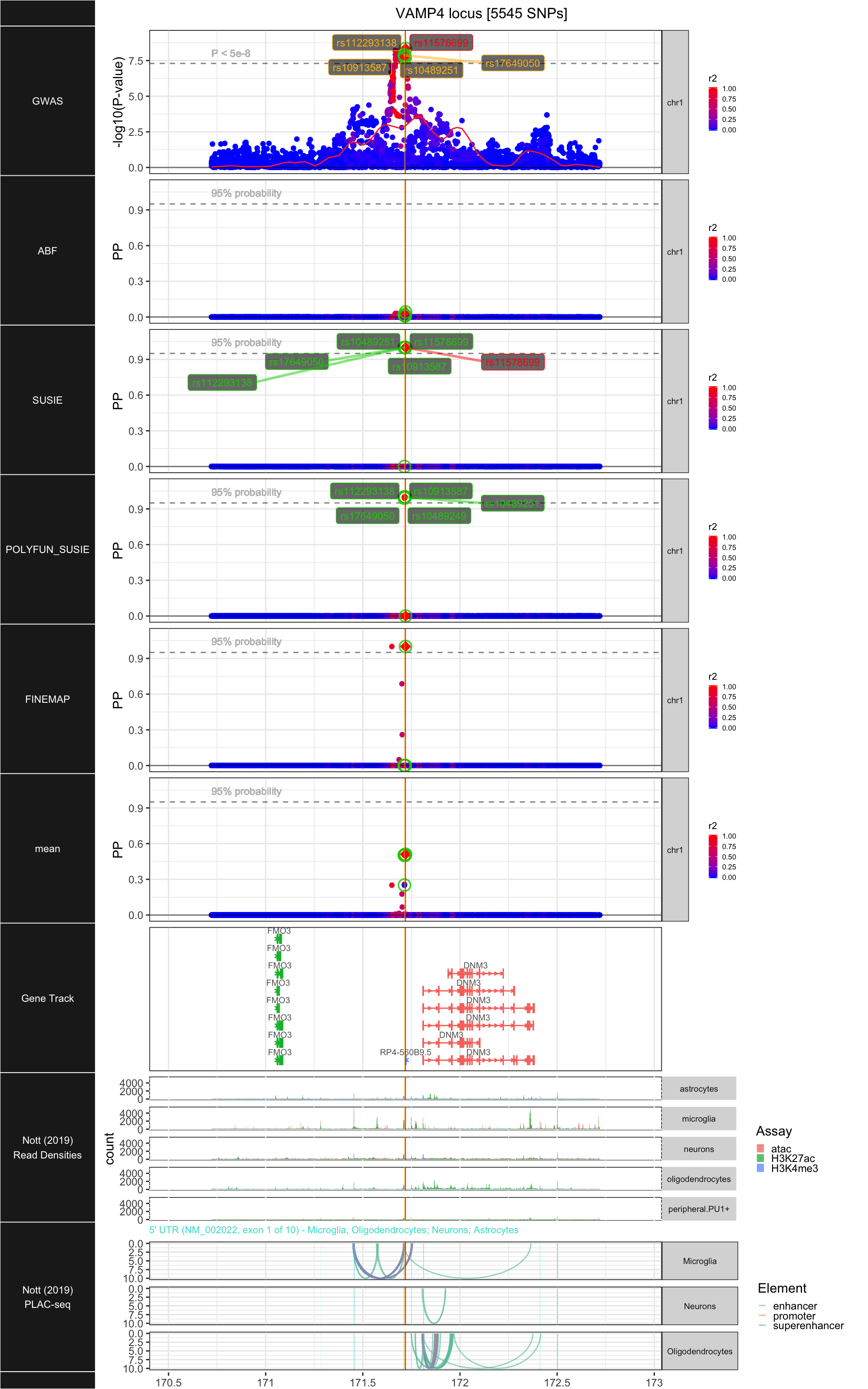
## ### VAMP4
## 
##
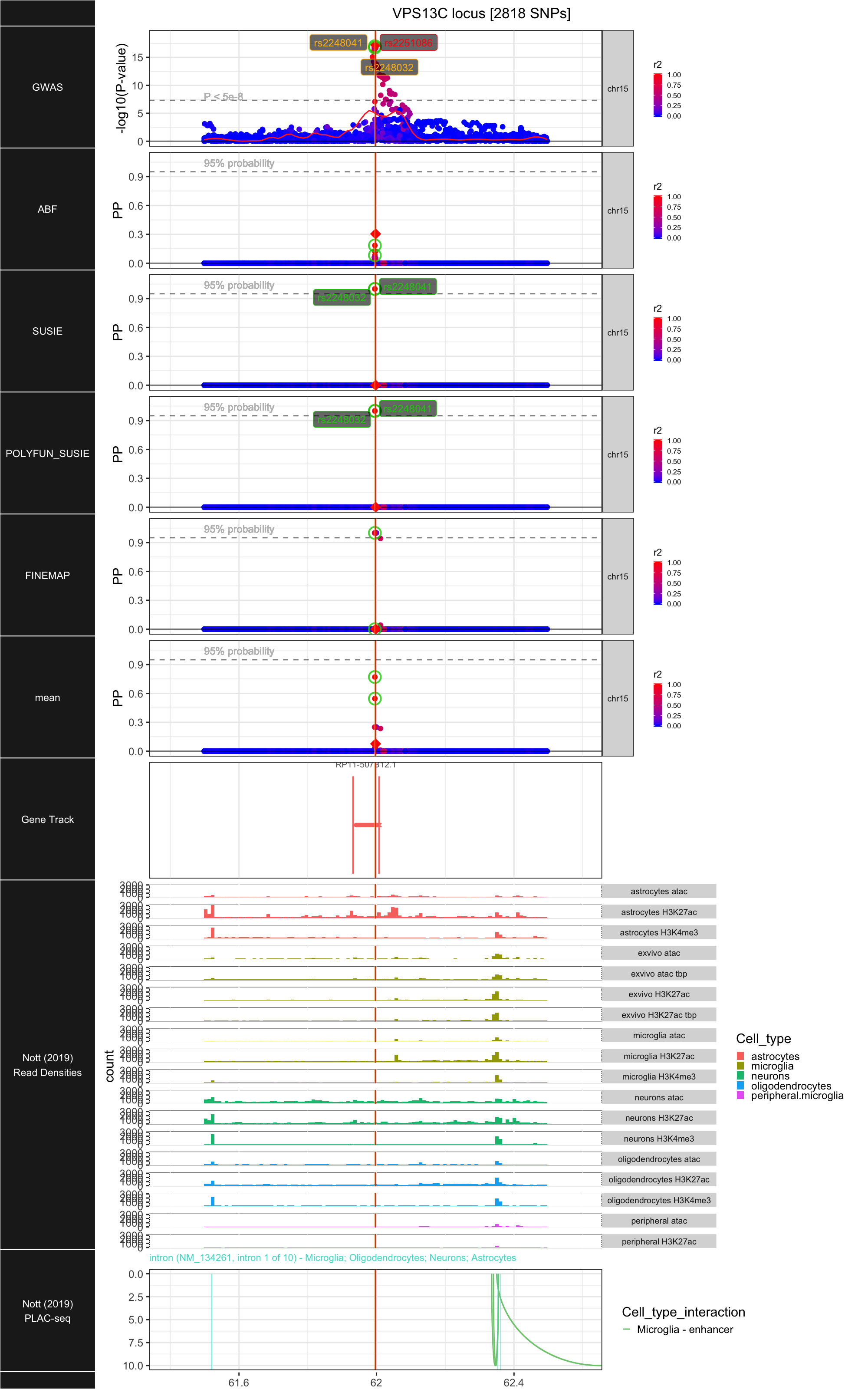
## ### VPS13C
## 
##
## ### WNT3
## 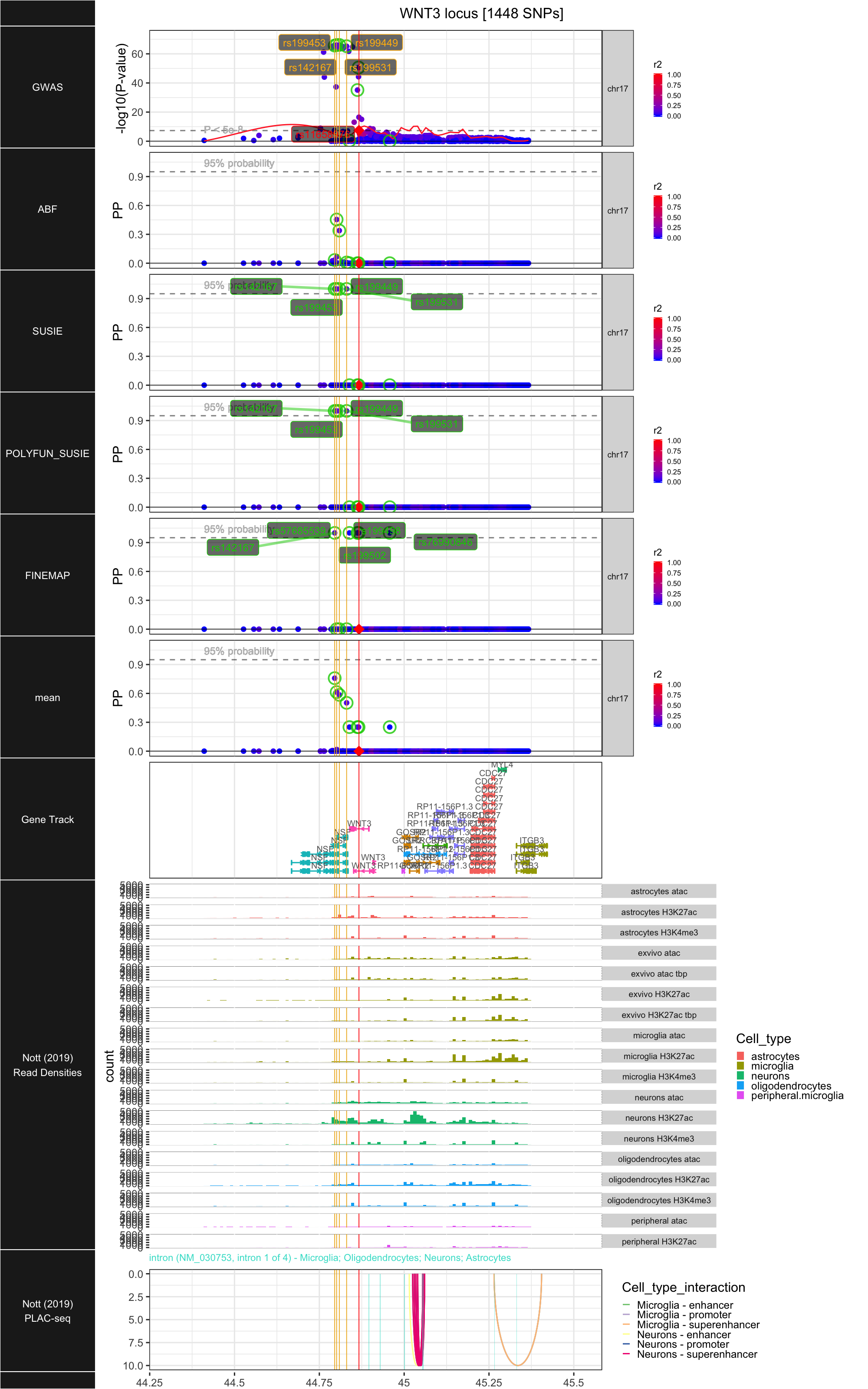## R version 3.6.1 (2019-07-05)
## Platform: x86_64-apple-darwin15.6.0 (64-bit)
## Running under: macOS Mojave 10.14.5
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] bsselectR_0.1.0 stringr_1.4.0 dplyr_0.8.3
##
## loaded via a namespace (and not attached):
## [1] Rcpp_1.0.3 crayon_1.3.4 digest_0.6.23 assertthat_0.2.1
## [5] R6_2.4.1 magrittr_1.5 evaluate_0.14 pillar_1.4.3
## [9] rlang_0.4.2 stringi_1.4.5 rmarkdown_2.0 tools_3.6.1
## [13] htmlwidgets_1.5.1 glue_1.3.1 purrr_0.3.3 xfun_0.12
## [17] yaml_2.2.0 compiler_3.6.1 pkgconfig_2.0.3 htmltools_0.4.0
## [21] tidyselect_0.2.5 knitr_1.26 tibble_2.1.3## [1] "susieR 0.8.0"